Apetala 2
| AP2 domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
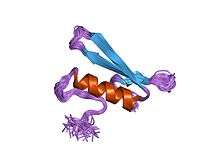 Structure of the GCC-box binding domain in complex with DNA.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | AP2 | ||||||||
| Pfam | PF00847 | ||||||||
| Pfam clan | CL0081 | ||||||||
| InterPro | IPR001471 | ||||||||
| SMART | SM00380 | ||||||||
| PROSITE | PS51032 | ||||||||
| SCOP | 3gcc | ||||||||
| SUPERFAMILY | 3gcc | ||||||||
| |||||||||
| Apetala 2 | |
|---|---|
| Identifiers | |
| Symbol | AP2 |
| Entrez | 829845 |
| UniProt | P47927 |
Apetala 2 (AP2) is a gene coding for a member of a large family of transcription factors, the AP2/EREBP family. In Arabidopsis thaliana which plays a role in the ABC model of flower development.[2] It was originally thought that this family of proteins was plant-specific; however, recent studies have shown that apicomplexans, including the causative agent of malaria, Plasmodium falciparum encode a related set of transcription factors, called the ApiAP2 family.[3]
In the A. thaliana transcription factor RAV1 the N-terminal AP2 domain binds 5'-CAACA-3' sequence, while the C-terminal highly conserved B3 domain binds 5'-CACCTG-3' sequence.[4]
Structure and Biological Content of AP2
Through recent studies, Apetala 2 is found to have a major role in hormone regulation,specific in flowers and plants, such as the AGAMOUS.[5] The study that determined this, done by Ogawa, was created to clarify the relationship between Apetala 2 and AtEBP in gene expression. The results showed that over-expression of AtEBP caused upregulation of AP2 expression in leaves which suggested that the N-terminal region is not required to produce AP2-like phenotypes.[6] AP2 also makes up another compound called ANT, which is composed of two AP2 domains homologous with the DNA binding domain of ethylene response element binding proteins.[7] Another study by Maes, T. titled Petunia Ap2-like genes and their role in flower and seed development, discovered three AP2-like proteins from petunia and by studying their expression patterns in situ hybridization: PhAP2A, PhAP2B, and PhAP2C. PhAP2A was found to have extremely similar functions of AP2 in A. thaliana and has an almost exact gene sequence. PhAP2B and PhAP2C encode for AP2-like proteins that belong to a different subgroup of the AP2 family of transcription factors and exhibit very different expression patterns during flower development compared to PhAP2A.[8]
Associations of AP2 with Chemical Compounds
Apetala 2 mutations cause changes in the ratio of hexose to sucrose during seed development, opening the possibility that AP2 may control seed mass through its effects on sugar metabolism.[9] As a protein, it regulates the amount of sugars in the system and is involved in transportation, shaping, and signaling. Another study showed that analyzed the functionality of DBF1 in abiotic stress responses and found that Arabidopsis plants over-expressing DBF1 were more tolerant to osmotic stress than control plants.[10] DBF1 is the binding factor that is found in the helping Apetala 2 carry out transcription factors.
Diseases
DNA is constantly subject to mutations, which can cause a complete shift in function of the protein due to the malformed protein, causing diseases in some cases. In an AP2 study for instance, an ERF/AP2-type transcription factor was isolated by differential-display reverse transcription-PCR, which induced a hypersensitive response in the leaves.[11] The Arabidopsis CBF gene family is composed of three genes encoding AP2domain-containing proteins, which are all regulated by low temperature to be able to carry out gene expression, but not by abscisic acid or dehydration.[12]
References
- ↑ Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M (September 1998). "A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA". EMBO J. 17 (18): 5484–96. doi:10.1093/emboj/17.18.5484. PMC 1170874
 . PMID 9736626.
. PMID 9736626. - ↑ Riechmann JL, Meyerowitz EM (1998). "The AP2/EREBP family of plant transcription factors". Biol. Chem. 379 (6): 633–46. doi:10.1515/bchm.1998.379.6.633. PMID 9687012.
- ↑ Balaji S, Babu MM, Iyer LM, Aravind L (2005). "Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains". Nucleic Acids Research. 33 (13): 3994–4006. doi:10.1093/nar/gki709. PMC 1178005
 . PMID 16040597.
. PMID 16040597. - ↑ Kagaya Y, Ohmiya K, Hattori T (1999). "RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants". Nucleic Acids Res. 27 (2): 470–8. doi:10.1093/nar/27.2.470. PMC 148202
 . PMID 9862967.
. PMID 9862967. - ↑ Mutual Regulation of Arabidopsis thaliana Ethylene-responsive Element Binding Protein and a Plant Floral Homeotic Gene, APETALA2. Ogawa, T., Uchimiya, H., Kawai-Yamada, M. Ann. Bot. (2007)
- ↑ Functional domains of the floral regulator AGAMOUS: characterization of the DNA binding domain and analysis of dominant negative mutations. Mizukami, Y., Huang, H., Tudor, M., Hu, Y., Ma, H. Plant Cell (1996)
- ↑ The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Klucher, K.M., Chow, H., Reiser, L., Fischer, R.L. Plant Cell (1996)
- ↑ Petunia Ap2-like genes and their role in flower and seed development. Maes, T., Van de Steene, N., Zethof, J., Karimi, M., D'Hauw, M., Mares, G., Van Montagu, M., Gerats, T. Plant Cell (2001)
- ↑ Control of seed mass by APETALA2. Ohto, M.A., Fischer, R.L., Goldberg, R.B., Nakamura, K., Harada, J.J. Proc. Natl. Acad. Sci. U.S.A. (2005)
- ↑ Maize DBF1-interactor protein 1 containing an R3H domain is a potential regulator of DBF1 activity in stress responses. Saleh, A., Lumbreras, V., Lopez, C., Kizis, E.D., Pagès, M. Plant J. (2006)
- ↑ The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Yi, S.Y., Kim, J.H., Joung, Y.H., Lee, S., Kim, W.T., Yu, S.H., Choi, D. Plant Physiol. (2004)
- ↑ The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression Is regulated by low temperature but not by abscisic acid or dehydration. Medina, J., Bargues, M., Terol, J., Pérez-Alonso, M., Salinas, J. Plant Physiol. (1999)
External links
- APETALA2 protein, Arabidopsis at the US National Library of Medicine Medical Subject Headings (MeSH)
- AP2 family, ERF family , RAV family At PlantTFDB: Plant Transcription Factor Database