Arylsulfatase B
| View/Edit Human | View/Edit Mouse |
| arylsulfatase B | |
|---|---|
 | |
| Identifiers | |
| Symbol | ARSB |
| Entrez | 411 |
| HUGO | 714 |
| OMIM | 253200 |
| RefSeq | NM_000046 |
| UniProt | P15848 |
| Other data | |
| EC number | 3.1.6.12 |
| Locus | Chr. 5 p11-q13 |
Arylsulfatase B (N-acetylgalactosamine-4-sulfatase, chondroitinsulfatase, chondroitinase, acetylgalactosamine 4-sulfatase, N-acetylgalactosamine 4-sulfate sulfohydrolase, EC 3.1.6.12) is an enzyme associated with mucopolysaccharidosis VI (Maroteaux–Lamy syndrome).
Arylsulfatase B is among a group of arylsulfatase enzymes present in the lysosomes of the liver, pancreas, and kidneys of animals. The purpose of the enzyme is to hydrolyze sulfates in the body. ARSB does this by breaking down glycosaminoglycans(GAGs), which are large sugar molecules in the body. ARSB targets two GAGs in particular: dermatan sulfate and chondroitin sulfate.[5]
Over 130 mutations to ARSB have been found, each leading to a deficiency in the body. In most cases, the mutation occurs on a single nucleotide in the sequence. An arylsulfatase B deficiency can lead to an accumulation of GAGs in lysosomes,[5] which in turn can lead to mucopolysaccharidosis VI.
Used as a pharmaceutical drug, the enzyme is known under the International Nonproprietary Name galsulfase and marketed as Naglazyme.[6]
Structure
The primary structure of Escherichia coli arylsulfatase B contains a primary sequence of 502 amino acids. Its secondary structure is quite complex, containing numerous alpha helices (20 total containing 138 residues) and beta sheets (21 strands total containing 87 residues).[4] The functional enzyme is believed to be a homo tetramer. Due to the complexity of arylsulfatase B's secondary structure, many hydrophobic and hydrophilic regions are present, as demonstrated by the Kyte-Doolittle hydropathy plot:
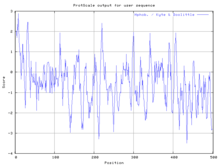
Additional structural data is shown in Ramachandran analysis plots at: http://www.rcsb.org/pdb/images/3ED4_ram_m_500.pdf.
Role in cystic fibrosis
Expression and activity of ARSB were found to be related to the function of cystic fibrosis transmembrane conductance regulator (CFTR), the membrane channel deficient in cystic fibrosis. Measurements in cystic fibrosis cell line IB3 and its derivative cell line C38, which has a functional CFTR, showed increased ARSB activity and expression in the C38 line.[7]
Role in malignancy
ARSB has been studied in a variety of cancers. Cultured normal mammary epithelial and myoepithelial cells had significantly higher ARSB activity than cultured malignant mammary cells.[8] Immunohistochemistry in the colon showed decreased membrane ARSB staining in colon cancer compared to normal colon, as well as in higher grade malignancies.[9] ARSB activity was lower in malignant than normal prostate tissue, and immunostaining of prostate tissue microarrays showed not only decreasing ARSB staining in prostate cancer tissue of a higher Gleason score, but also lower staining in patients with recurrent compared to non-recurrent cancer. ARSB staining was a greater predictor of recurrence than Prostate-specific antigen (PSA) test, indicating possible future role of ARSB as a prognostic biomarker of prostate cancer.[10] Further evidence of ARSB as a tumor suppressor was determined by molecular studies in cell cultures where ARSB was silenced by siRNA. The studies showed that decrease of ARSB leads to increase in free galectin-3, which attaches more strongly to less sulfated chondroitin 4-sulfate. Galectin-3 then acts on transcription factors AP-1 to increase expression of chondroitin sulfate proteoglycan versican and SP-1 to increase expression of WNT9A.[11][12]
Extra-lysosomal localization
Although primarily a lysosomal enzyme, ARSB was also found to localize at the cell membrane of hepatocytes, sinusoidal endothelial cells, and Kupffer cells in the liver, as well as in the apical membranes of normal and malignant colonic and prostatic epithelial cells, by immunohistochemistry and immunofluorescence studies. Membrane immunostaining in the colon and prostate was lower in malignant than in normal tissue and also was lower in higher grade malignancies.[9][10][13] ARSB activity assay in the membrane and cytosol fractions of cultured bronchial epithelial cells showed that the activity was several-fold greater in the membrane fraction.[14]
Read More: http://www.atsjournals.org/doi/full/10.1165/rcmb.2008-0482OC#.VlSq-17h0bA
Read More: http://www.atsjournals.org/doi/full/10.1165/rcmb.2008-0482OC#.VlSq-17h0bA
See also
References
- ↑ "Diseases that are genetically associated with ARSB view/edit references on wikidata".
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- 1 2 PDB: 3ED4; Patskovsky Y, Ozyurt S, Gilmore M, Chang S, Bain K, Wasserman S, Koss J, Sauder MJ, Burley SK, Almo SC (2010). "Crystal structure of putative arylsulfatase from Escherichia coli". To be Published. doi:10.2210/pdb3ed4/pdb.
- 1 2 U.S. National Library of Medicine. "ARSB", Genetics Home Resource, November 7, 2010, Retrieved November 22, 2010
- ↑ Kim KH, et al. (2008). "Successful management of difficult infusion-associated reactions in a young patient with mucopolysaccharidosis type VI receiving recombinant human arylsulfatase B (galsulfase [Naglazyme])". Pediatrics. 121: e714–7. doi:10.1542/peds.2007-0665. PMID 18250117.
- ↑ Sumit Bhattacharyya, Dwight Look, Joanne K. Tobacman. Increased arylsulfatase B activity in cystic fibrosis cells following correction of CFTR. Clinica Chimica Acta, Volume 380, Issues 1–2, 1 May 2007, Pages 122–12
- ↑ Sumit Bhattacharyyaa, Joanne K. Tobacman. Steroid sulfatase, arylsulfatases A and B, galactose-6-sulfatase, and iduronate sulfatase in mammary cells and effects of sulfated and non-sulfated estrogens on sulfatase activity. The Journal of Steroid Biochemistry and Molecular Biology. Volume 103, Issue 1, January 2007, Pages 20–34
- 1 2 Sanjiv V. Prabhu, Sumit Bhattacharyya, Grace Guzman-Hartman, Virgilia Macias, André Kajdacsy-Balla, Joanne K. Tobacman. Extra-Lysosomal Localization of Arylsulfatase B in Human Colonic Epithelium. J Histochem Cytochem March 2011 vol. 59 no. 3 328-335 doi: 10.1369/0022155410395511
- 1 2 L Feferman, S Bhattacharyya, R Deaton, P Gann, G Guzman, A Kajdacsy-Balla, and JK Tobacman. Arylsulfatase B (N-acetylgalactosamine-4-sulfatase): potential role as a biomarker in prostate cancer. Prostate Cancer and Prostatic Disease (2013), 1–8
- ↑ S Bhattacharyya, L Feferman and JK Tobacman. Arylsulfatase B regulates versican expression by galectin-3 and AP-1 mediated transcriptional effects. Oncogene (2013), 1–10
- ↑ Sumit Bhattacharyya, Leo Feferman and Joanne K. Tobacman. Increased Expression of Colonic Wnt9a through Sp-1 mediated Transcriptional Effects Involving Arylsulfatase B, Chondroitin-4-Sulfate, and Galectin-3. J. Biol. Chem. published online April 28, 2014
- ↑ Keiko Mitsunaga-Nakatsubo, Shinichiro Kusunoki, Hayato Kawakami, Koji Akasaka, Yoshihiro Akimoto. Cell-surface arylsulfatase A and B on sinusoidal endothelial cells, hepatocytes, and Kupffer cells in mammalian livers. Med Mol Morphol (2009) 42:63–69 DOI 10.1007/s00795-009-0447-x
- ↑ Sumit Bhattacharyya, Kemal Solakyildirim, Zhenqing Zhang, Mei Ling Chen, Robert J. Linhardt, and Joanne K. Tobacman. Cell-Bound IL-8 Increases in Bronchial Epithelial Cells after Arylsulfatase B Silencing due to Sequestration with Chondroitin-4-Sulfate. American Journal of Respiratory Cell and Molecular Biology, Vol. 42, No. 1 (2010), pp. 51-61. doi: 10.1165/rcmb.2008-0482OC
External links
- Medical Dictionary, Arylsulfatase B function.
- Protein Data Base (PDB), Arylsulfatase structure.
- Genetics Home Reference, Arylsulfatase B function.