BCKDK
| View/Edit Human | View/Edit Mouse |
Branched chain ketoacid dehydrogenase kinase (BCKDK) is an enzyme encoded by the BCKDK gene on chromosome 16. This enzyme is part of the mitochondrial protein kinases family and it is a regulator of the valine, leucine, and isoleucine catabolic pathways.[3] BCKDK is found in the mitochondrial matrix and the prevalence of it depends on the type of cell. Liver cells tend to have the lowest concentration of BCKDK, whereas skeletal muscle cells have the highest amount.[4][5] Abnormal activity of this enzyme often leads to diseases such as maple syrup urine disease and cachexia.
Structure
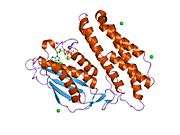
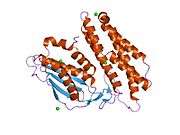
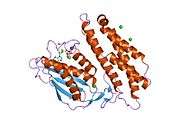
BCKDK’s structure consists of a characteristic nucleotide-binding domain along with a four-helix bundle domain similar to certain aspects of protein histidine kinases, which are involved in two-component signal transduction systems. BCKDK is also a dimer with a Leu389 residue located between the dimers and this dimerization is seen to be essential for its kinase activity and protein stability.[6] Moreover, it is made up of 382 amino acids and has a molecular weight of 43 kDa.[4] The gene BCKDK is located at 16p11.2, has an exon count of 11, and it lacks a TATA-box and an initiator element.[3][5]
Function
BCKDK regulates the activity of branched-chain α-ketoacid dehydrogenase complex (BCKD) through phosphorylation and inactivation. This inactivation results in increased branched-chain amino acids (BCAA), which is seen to reduce oxidative stress; however, having too much BCAA has been proven to be toxic to humans. Therefore, BCKDK is a vital tool to assist with BCAA homeostasis.[7][8] As stated earlier, BCKDK concentrations vary depending on the type of tissue that is observed, whereas BCKD’s concentration is the same in any tissue. Although BCKD concentration is constant, the amount of BCKDK determines the activity of the dehydrogenase complex. Since liver tissue is seen to have the lowest concentration of BCKDK, the activity of BCKD is seen to be the highest, delineating the fact that the BCKD kinase inversely affects the BCKD activity.[5]
Clinical Significance
Abnormalities in BCKD activity often leads to pathological conditions which is why BCKDK is needed to regulate it. Often, mutations in the BCKDK gene occur creating the deviation in BCKD behavior. Exceedingly high BCKD complex activity increases branched-chain amino acid catabolism and protein degradation in skeletal muscle, which is a distinctive feature for cachexia. Deficiencies in BCKD activity have been the main cause in the rare metabolism maple syrup urine disease that can lead to mental retardation, brain edema, seizures, coma, and death if not treated correctly by lifelong limitation of branched-chain amino acid intake.[5] Because BCKDK regulates BCKD which in turn catalyzes BCAA, BCKDK is one of the factors that determines the concentration of BCAA levels. High BCAA levels can lead to insulin resistance and can be a potential marker for type 2 diabetes.[8] The amalgamation of BCAA can also lead to congenial heart diseases and heart failure. Furthermore, low levels of BCAA have been described as a cause of comorbid intellectual disability, autism, and epilepsy.[6]
Interactions
BCKDK has been seen to interact with:
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- 1 2 "Entrez Gene: BCKDK branched chain ketoacid dehydrogenase kinase".
- 1 2 3 Popov KM, Zhao Y, Shimomura Y, Kuntz MJ, Harris RA (Jul 1992). "Branched-chain alpha-ketoacid dehydrogenase kinase. Molecular cloning, expression, and sequence similarity with histidine protein kinases". The Journal of Biological Chemistry. 267 (19): 13127–30. PMID 1377677.
- 1 2 3 4 Muller EA, Danner DJ (Oct 2004). "Tissue-specific translation of murine branched-chain alpha-ketoacid dehydrogenase kinase mRNA is dependent upon an upstream open reading frame in the 5'-untranslated region". The Journal of Biological Chemistry. 279 (43): 44645–55. doi:10.1074/jbc.M406550200. PMID 15302860.
- 1 2 García-Cazorla A, Oyarzabal A, Fort J, Robles C, Castejón E, Ruiz-Sala P, Bodoy S, Merinero B, Lopez-Sala A, Dopazo J, Nunes V, Ugarte M, Artuch R, Palacín M, Rodríguez-Pombo P, Alcaide P, Navarrete R, Sanz P, Font-Llitjós M, Vilaseca MA, Ormaizabal A, Pristoupilova A, Agulló SB (Apr 2014). "Two novel mutations in the BCKDK (branched-chain keto-acid dehydrogenase kinase) gene are responsible for a neurobehavioral deficit in two pediatric unrelated patients". Human Mutation. 35 (4): 470–7. doi:10.1002/humu.22513. PMID 24449431.
- ↑ Tso SC, Gui WJ, Wu CY, Chuang JL, Qi X, Skvora KJ, Dork K, Wallace AL, Morlock LK, Lee BH, Hutson SM, Strom SC, Williams NS, Tambar UK, Wynn RM, Chuang DT (Jul 2014). "Benzothiophene carboxylate derivatives as novel allosteric inhibitors of branched-chain α-ketoacid dehydrogenase kinase". The Journal of Biological Chemistry. 289 (30): 20583–93. doi:10.1074/jbc.M114.569251. PMID 24895126.
- 1 2 Tso SC, Qi X, Gui WJ, Chuang JL, Morlock LK, Wallace AL, Ahmed K, Laxman S, Campeau PM, Lee BH, Hutson SM, Tu BP, Williams NS, Tambar UK, Wynn RM, Chuang DT (Jun 2013). "Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain α-ketoacid dehydrogenase kinase". Proceedings of the National Academy of Sciences of the United States of America. 110 (24): 9728–33. doi:10.1073/pnas.1303220110. PMID 23716694.
Further reading
- Popov KM, Zhao Y, Shimomura Y, Kuntz MJ, Harris RA (Jul 1992). "Branched-chain alpha-ketoacid dehydrogenase kinase. Molecular cloning, expression, and sequence similarity with histidine protein kinases". The Journal of Biological Chemistry. 267 (19): 13127–30. PMID 1377677.
- Chang CF, Chou HT, Chuang JL, Chuang DT, Huang TH (May 2002). "Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex". The Journal of Biological Chemistry. 277 (18): 15865–73. doi:10.1074/jbc.M110952200. PMID 11839747.
- Xu YC, Wu RF, Gu Y, Yang YS, Yang MC, Nwariaku FE, Terada LS (Aug 2002). "Involvement of TRAF4 in oxidative activation of c-Jun N-terminal kinase". The Journal of Biological Chemistry. 277 (31): 28051–7. doi:10.1074/jbc.M202665200. PMID 12023963.
- Starcevic M, Dell'Angelica EC (Jul 2004). "Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1)". The Journal of Biological Chemistry. 279 (27): 28393–401. doi:10.1074/jbc.M402513200. PMID 15102850.
- Wynn RM, Kato M, Machius M, Chuang JL, Li J, Tomchick DR, Chuang DT (Dec 2004). "Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation". Structure. 12 (12): 2185–96. doi:10.1016/j.str.2004.09.013. PMID 15576032.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (Oct 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. doi:10.1038/nature04209. PMID 16189514.
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D (2007). "Large-scale mapping of human protein-protein interactions by mass spectrometry". Molecular Systems Biology. 3 (1): 89. doi:10.1038/msb4100134. PMC 1847948
 . PMID 17353931.
. PMID 17353931.
External links
- BCKDK human gene location in the UCSC Genome Browser.
- BCKDK human gene details in the UCSC Genome Browser.