Neuronal lineage marker
A Neuronal lineage marker is an endogenous tag that is expressed in different cells along neurogenesis and differentiated cells such as neurons. It allows detection and identification of cells by using different techniques. A neuronal lineage marker can be either DNA, mRNA or RNA expressed in a cell of interest. It can also be a protein tag, as a partial protein, a protein or an epitope that discriminates between different cell types or different states of a common cell. An ideal marker is specific to a given cell type in normal conditions and/or during injury. Cell markers are very valuable tools for examining the function of cells in normal conditions as well as during disease. The discovery of various proteins specific to certain cells led to the production of cell-type-specific antibodies that have been used to identify cells.[1]
The techniques used for its detection can be immunohistochemistry, immunocytochemistry, methods that utilize transcriptional modulators and site-specific recombinases to label specific neuronal population,[2] in situ hybridization or fluorescence in situ hybridization (FISH).[3] A neuronal lineage marker can be a neuronal antigen that is recognized by an autoantibody for example Hu, which is highly restricted to neuronal nuclei. By immunohistochemistry, anti-Hu stains the nuclei of neurons.[4] To localize mRNA in brain tissue, one can use a fragment of DNA or RNA as a neuronal lineage marker, a hybridization probe that detects the presence of nucleotide sequences that are complementary to the sequence in the probe. This technique is known as in situ hybridization. Its application have been carried out in all different tissues, but particularly useful in neuroscience. Using this technique, it is possible to locate gene expression to specific cell types in specific regions and observe how changes in this distribution occur throughout the development and correlate with the behavioral manipulations.[5]

Although immunohistochemistry is the staple methodology for identifying neuronal cell types, since it is relatively low in cost and a wide range of immunohistochemical markers are available to help distinguish the phenotype of cells in the brain, sometimes it is time-consuming to produce a good antibody.[6] Therefore, one of the most convenient methods for the rapid assessment of the expression of a cloned ion channel could be in situ hybridization histochemistry.
After cells are isolated from tissue or differentiated from pluripotent precursors, the resulting population needs to be characterized to confirm whether the target population has been obtained. Depending on the goal of a particular study, one can use neural stem cells markers, neural progenitor cell markers, neuron markers or PNS neuronal markers.
History
The study of the nervous system dates back to ancient Egypt but only in the ninetieth century it became more detailed. With the invention of the microscope and a technique of staining developed by Camillo Golgi, it was possible to study individual neurons. This scientist started to impregnate nervous tissue with metal, as silver. The reaction consists in fixing particles of silver chromate to the neurilemma, and resulted in a stark black deposit in the soma, axon and dendrites of the neuron. Thus, it was possible to identify different types of neurons, as Golgi Cell, Golgi I and Golgi II.[7] In 1885 there was a German medical researcher called Franz Nissl who developed another staining technique now known by Nissl staining. This technique is slightly different from Golgi staining since it stains the cell body and the endoplasmic reticulum.[8] In 1887, a Spanish scientist called Santiago Ramon y Cajal learned the staining technique with Golgi and started his famous work of neuroanatomy.[9] With this technique he made an extensive study of several areas of the brain and in different species. He also described very precisely the purkinje cells, the chick cerebellum and the neuronal circuit of the rodent hippocampus. In 1941 Dr. Albert Coons used for the first time a revolutionary technique that uses the principle of antibodies binding specifically to antigens in the tissues. He created an immunoflorescent technique for labelling the antibodies.[10] This technique continues to be widely used in neuroscience studies for identifying different structures. The most important neural markers used nowadays are the GFAB, Nestin, NeuroD antibodies and others.[11] For the past years there are still creating new neural markers for immunocytochemistry or/and immunohistochemistry. In 1953 Heinrich Klüver invented a new staining technique called, Luxol Fast Blue stain or LFB, and with this technique it’s possible to detect demyelination in the central nervous system. Myelin sheath will be stained blue, but other structures will be stained as well. The next revolutionary technique was invented in 1969 by an American scientist called Joseph G. Gall.[12] This technique is called in situ Hybridization and it is used in a large variety of studies but mainly used in developmental biology. With this technique it is possible to mark some genes expressed in determined areas of the animal. In neurobiology, it's very useful for understanding the formation of the nervous system.
Techniques
In situ hybridization
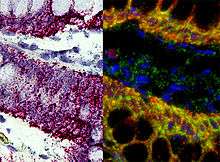
This is one of the most powerful techniques to mark cells. This method consists of hybridizing a labeled complementary DNA or RNA strand to a specific DNA or RNA in the tissue. By doing this hybridization we will be able to reveal the location of a specific mRNA, giving us information about the physiological process of organization, regulation and function of the genes.[13]
Using this technique we can now know what are the genes and proteins that are behind a certain process, like the formation of the neural crest, or a specific behavior; and what is the location of that same genes. We can also see how changes in the distribution of these genes can affect the development of a tissue, and correlate it with behavioral manipulations.[13] Some examples are the use of, digoxigenin- or fluorophore-conjugated oligo- nucleotide probes, for the detection of localized mRNAs in dendrites, spines, axons, and growth cones of cultured neurons; or digoxigenin-labeled RNA probes and fluorescence tyramide amplification for the detection of less abundant mRNAs localized to dendrites in vivo.[14] These examples use FISH (Fluorescent in situ hybridization). With this technique we can understand the physiological processes and neurological diseases.

Immunohistochemistry
Immunohistochemistry is a technique that uses antibodies with fluorescent staining tags that target a specific antigen present in a certain protein. This high specificity allows us to localize the peptidergic and classical transmitter compounds, their synthetic enzymes and other cell specific antigen in neuronal tissiue.
An example of the application of this technique in neuroscience is the immunolabeling of antigens like NGF-Inducible Large External glycoprotein (NILE-GF), choline acetyltransferase, parvalbumin, and neurofilament protein.[15] All of these antigens are present in specific neuronal cell types. With these we can define anatomical circuits with a high degree of resolution, and understand the role of some proteins and cells in the nervous system, as well as the location of that same proteins and cells.
Although this is a very potent technique there are some drawbacks. The procedure has a nonquantifiable nature and has the occurrence of both false positives and false negatives.[16]
Immunocytochemistry
Immunocytochemistry uses the same method that immunohistochemistry, but with the difference that this technique is used in isolated cells in culture, and the other is in tissues. The results are the same but with more resolution, once we are looking to one cell only.
Lineage markers
Neural stem cells markers
Neural stem cells are an example of somatic stem cell found in various tissues, both during development and in the adult. They have two fundamental characteristics: they are self-renewing and upon terminal division and differentiation, they can give rise to the full range of cells classes within the relevant tissue. Hence, a neural stem cell can give rise to another neural stem cell, or to any of the differentiated cell types found in the central and peripheral nervous systems (inhibitory and excitatory neurons, astrocytes and oligodendrocytes).[17]
The standard method of isolating neural stem cells in vitro is with the neurosphere culture system, the method originally used to identify NSCs. After some proliferation, the cells are either induced to differentiate by withdrawing the mitogens or by exposing the cells to another factor that induces some of the cells to develop into different lineages.[18] Cellular fates are analysed by staining with antibodies directed against antigens specific for astrocytes, oligodendrocytes, and neurons. In some cases, cells are plated at low density and monitored to determine if a single cell can give rise to the three phenotypes.[19] Immunomagnetic cell separation strategies using antibodies directed against cell surface markers present on stem cells, progenitors and mature CNS cells have been applied to the study of NSCs. Other non-immunological methods have been used to identify populations of cells from normal and tumorigenic CNS tissues, which demonstrate some of the in vitro properties of stem cells, including high aldehyde dehydrogenase (ALDH) enzyme activity. ALDH cells from embryonic rat and mouse CNS have been isolated and shown to have the ability to generate neurospheres, neurons, astrocytes and oligodendrocytes in vitro, as well as neurons in vivo when transplanted into the adult mouse cerebral cortex.[18] Once a stem cell divides asymmetrically, the more mature progenitor is born and migrates to regions of differentiation. As the progenitor migrates, it matures further until it reaches a site where it stops and either becomes quiescent or fully differentiates into a functioning cell. The major obstacle to identifying and discovering markers that define a stem cell is that the most primitive cells are probably in a quiescent state and do not express many unique antigens. Thus, as with other fields like haematopoiesis, a combination of positive and negative markers will be required to better define the central nervous system stem cell.[19] Nonetheless, changes in the expression levels of specific molecules can be used to indicate the presence of neural stem cells in studies focused on further differentiation toward specific neural lineages. Usual markers used for neural stem cells include Nestin and SOX2. Although Nestin it is expressed predominantly in stem cells of the central nervous system (CNS), its expression is absent from nearly all mature CNS cells, thus it is an efficient marker for neural stem cells.[20] During neurogenesis, Sox2 is expressed throughout developing cells in the neural tube as well as in proliferating CNS progenitors, hence is thought to be centrally important for neural stem cell proliferation and differentiation.[20] In addition to intracellular molecules, products are available to study proteins which are expressed at the cell surface, including ABCG2, FGF R4, and Frizzled-9.
Differentiation markers
The differentiation of neural stem cells is controlled, in a context-dependent manner, by intrinsic factors and extracellular signalling molecules that act as positive or negative regulators that can be used as markers.[21]
Neural progenitor markers
A neural progenitor cell is distinct from a neural stem cell since it is incapable of continuous self-renewal and usually has the capacity to give rise to only one class of differentiated progeny. They are tripotent cells which can give rise to neurons, astrocytes and oligodendrocytes. An oligodendroglial progenitor cell, for example, gives rise to oligodendrocytes until its mitotic capacity is exhausted.[17] Some neural progenitor markers are capable of tracking cells as they undergo expansion and differentiation from rosettes to neurons. The neural rosette is the developmental signature of neuroprogenitors in cultures of differentiating embryonic stem cells; rosettes are radial arrangements of columnar cells that express many of the proteins expressed in neuroepithelial cells in the neural tube. It has been shown that cells within rosettes express multiple cell markers, including among others Nestin, NCAM and Musashi-1, a RNA-binding protein that is expressed in proliferating neural stems cells.[22] Neuroepithelial progenitors (NEP) are responsible for neurogenesis in the neural tube and also give rise to two other types of neural progenitor cell, radial glia and basal progenitors. Radial glia are the dominant progenitor cell type in the developing brain whereas basal progenitors are specifically located at the subventricular zone (SVZ) in the developing telencephalon. Although functional studies of radial glia are increasing, it is difficult to distinguish them from neuroprogenitors and astrocytes. Like neuroprogenitors, radial glia express intermediate filament proteins nestin as well as the transcription factor PAX6 that is expressed in some neuroprogenitors in the ventral half of the neural tube. Radial glia also express proteins characteristic of astrocytes, including the widely used glial fibrillar acidic protein (GFAP), among others. Cytological markers that might be unique to radial glia include modified forms of nestin identified by the RC1 and RC2 antibodies that recognize the murine antigens.[23]
Neuron markers
Markers can detect neurons in different stages of development from nuclear, cytoplasmic, membrane or perisynaptic products present in neurons. It is also possible to label specifically cholinergic, dopaminergic, serotonergic, GABAergic or glutamatergic neurons.[24] Pan neuron markers have multiple targets (somatic, nuclear, dendritic, spine and axonal proteins) and consequently label across all parts of the neuron, it is used for study neuronal morphology. Although there are specific markers that label particular regions of the neuron.[25]
Doublecortin (DCX) is a microtubule-associated protein that is widely expressed in the soma and leading processes of migrating neurons and in the axons of differentiation neurons. Its expression is downregulated with maturation [26]
Neuron-specific Class III β-tubulin (TuJ1) is present in newly generated immature postmitotic neurons and differentiated neurons and in some mitotically active neuronal precursors.[11]
Microtubule-associated protein 2 (MAP-2) is a cytoskeletal protein. Its expression is weak in neuronal precursors but it increases during neuron development process. In general, its expression is confined to neurons and reactive astrocytes.[15]
Neuron specific enolase (NSE), also called as gamma-enolase or enolase 2, is a cytosolic protein that is expressed in mature neurons. NSE levels increase along the neuron development reaching higher level in later stages. It can be expressed in glial cells during oligodendrocyte differentiation with the same levels that have been found in neuron culture, but is repressed when cells become mature. In pathological conditions was also reported that glial neoplasms and reactive glial cells expressed this marker.[15]
Calretinin is widely distributed in different neuronal populations of vertebrate retina, being a valuable marker for immature postmitotic neurons.[11]
Neuronal Nuclei antigen (NeuN) or Fox-3 is a nuclear protein present in postmitotic stage when start to differentiate into mature cells.[27] It allows to detect all the neuronal cell types except Purkinje cells, olfactory bulb mitral cells, retinal photoreceptor and dopaminergic neurons in the substantia nigra.[15]
Calbindin is expressed by cerebellar Purkinje cells and granule cells of hippocampus.[11] The reorganization and migration of calbindin-stained Purkinje neurons in rat cerebellum after peripheral nerve injury suggests that calbindin may be a marker for immature post-mitotic neurons, similar to calretinin.[28]
Tyrosine hydroxylase (TH) is an enzyme involved in the synthesis of dopamine and norepinephrine. Generally, it is used as a marker for dopaminergic neurons, but it can also be found in some forebrain neurons which make norepinephrine (which is the product of dopamine and the enzyme dopamine β-hydroxylase).[29]
Choline Acetyltransferase (ChAT) is expressed in cholinergic neurons of both the CNS and PNS. In the CNS, ChAT is expressed in motor neurons and pre-ganglionic autonomic neurons of the spinal cord, a subset of neurons in the neostriatum and in the basal forebrain. On the other hand, in PNS it is present in a small group of sympathetic neurons and in all parasympathetic neurons.[30]
GABA is a mature neuronal marker expressed in GABAergic interneurons (inhibitor neurons which are generally interneurons in the brain). GAD65/67 are two enzymes involved in GABA synthesis by GABAergic interneurons.[29]
Clinical research
Neuronal lineage markers can be used in clinical research to identify diseased cells and/or in repair process. Since selective degeneration of functional neurons is associated with the pathogenesis of neurodegenerative disorders, such as degeneration of midbrain dopaminergic neurons in Parkinson's disease, forebrain cholinergic neurons in Alzheimer's disease and cortical GABAergic neurons in schizophrenia, markers of neuronal cell phenotype are of particular interest because of their utility in understanding pathology of clinical disease.[31] There are two key markers in these studies: choline acetyltransferase and tyrosine hydroxylase. Choline acetyltransferase (ChAT) is an enzyme responsible for catalyzing the synthesis of acetylcholine, and is expressed in the majority of cholinergic neurons. Hence, ChAT immunoreactivity is used to detect cognitive decline in several neurodegenerative disorders.[15] In motor regions, sensory cortex and in the basal forebrain these immunolabeling has been applied to evaluate disruptions in cholinergic neurons of the ChAT fiber network and also for overall morphology.[32] The Tyrosine hydroxylase (TH) immunolabeling has been very useful for Parkinson's disease investigation. It is used to determine the quantity of dopaminergic cell loss in Parkinson’s patients.
Examples of neuronal lineage markers
| Cell types | Markers |
|---|---|
| Neural Stem Cell |
ABCG2; NeuroD1; ASCL1/Mash1; Noggin; Beta-catenin; Notch-1; Notch-2; Brg1 ; Nrf2 ; N-Cadherin; Nucleostemin; Calcitonin R; Numb; CD15/Lewis X; Otx2; CDCP1; Pax3; COUP-TF I/NR2F1; Pax6; CXCR4; PDGF R alpha; FABP7/B-FABP; PKC zeta; FABP 8/M-FABP; Prominin-2; FGFR2; ROR2; FGFR4; RUNX1/CBFA2; FoxD3; RXR alpha/NR2B1; Frizzled-9; sFRP-2; GATA-2; SLAIN 1; GCNF/NR6A1; SOX1; GFAP; SOX2; Glut1; SOX9; HOXB1; SOX11; ID2; SOX21; Meteorin; SSEA-1; MSX1; TRAF-4; Musashi-1; Vimentin; Musashi-2; ZIC1; Nestin |
| Neural Progenitor Markers |
A2B5; AP-2 Alpha; ATPase Na+/K+ transporting alpha 1; Activin RIIA; Brg1; CD168/RHAMM; CD4; Doublecortin/DCX; Frizzled 4/CD344; GAP43; Jagged1; Laminin; MSX1/HOX7; Mash1; Musashi-1; Nestin; Netrin-1; Netrin-4; Neuritin; NeuroD1; Neurofilament alpha-internexin/NF66; Notch1; Notch2; Notch3; Nucleostemin; Otx2; PAX3; S100B; SOX2; Semaphorin 3C; Semaphorin 6A; Semaphorin 6B; Semaphorin 7A; TROY/TNFRSF19; Tubulin βII; Tuj 1; Vimentin |
| Early Neuronal Markers |
ATH1/MATH1; ASH1/MASH1; HES5; HuC/Hu; HuD; Internexin α; L1 neural adhesion molecule; MAP1B/MAP5; MAP2A; MAP2B; Nerve Growth Factor Rec/NGFR; Nestin; NeuroD; Neurofilament L 68 kDa; Neuron Specific Enolase/NSE; NeuN; Nkx-2.2/NK-2; Noggin; Pax-6; PSA-NCAM; Tbr1; Tbr2; Tubulin βIII; TUC-4; Tyrosine hydroxylase/TH |
| Immature Neuron & Growth Cone Markers |
Calbindin; Calretinin; Collapsin Response Mediated Protein 1 /CRMP1; Collapsin Response Mediated Protein 2 /CRMP2; Collapsin Response Mediated Protein 5 /CRMP5; Contactin-1; Cysteine-rich motor neuron 1/CRIM1; c-Ret phosphor Serine 696; Doublecortin/DCX; Ephrin A2; Ephrin A4; Ephrin A5; Ephrin B1; Ephrin B2; GAP-43; HuC; HuD; Internexin alpha; Laminin-1; LINGO-1; MAP1B/MAP5; Mical-3; NAP-22; NGFR; Nestin; Netrin-1; Neuropilin; Plexin-A1; RanBPM; Semaphorin 3A; Semaphorin 3F; Semaphorin 4D; Slit2; Slit3; Staufen; Tbr 1; Tbr 2; Trk A; Tubulin βIII; TUC-4 |
| Differentiated Post-Mitotic Neuronal Cells |
NeuN; NF-L; NF-M; GAD; TH; PSD-95; Synaptophysin; VAMP; ZENON |
| Motor Neurons |
ChAT/choline acetyltransferase Chox10; En1; Even-skipped/Eve; Evx1; Evx2; Fibroblast growth factor-1/FGF1; HB9; Isl1; Isl2; Lim3; Nkx6; p75 neurotrophin receptor; REG2; Sim1; SMI32; Zfh1 |
| Perisynaptic |
4.1G; Acetylcholinesterase; Ack1; AMPA Receptor Binding Protein/ABP; ARG3.1; Arp2; E-Cadherin; N-Cadherin; Calcyon; Catenin alpha and beta; Caveolin; CHAPSYN-110/PSD93; Chromogranin A; Clathrin light chain; Cofilin; Complexin 1/CPLX1/Synaphin 2; Contactin-1; CRIPT; Cysteine String Protein/CSP; Dynamin 1; Dymanin 2; Flotillin-1; Fodrin; GRASP; GRIP1; Homer; Mint-1; Munc-18; NSF; PICK1; PSD-95; RAB4; Rabphillin 3A; SAD A; SAD B; SAP-102; SHANK1a; SNAP-25; Snapin; Spinophilin/Neurabin-1; Stargazin; Striatin; SYG-1; Synaptic Vesicle Protein 2A; Synaptic Vesicle Protein 2B; Synapsin 1; Synaptobrevin/VAMP; Synaptojanin 1; Synaptophysin; Synaptotagmin; synGAP; Synphilin-1; Syntaxin 1; Syntaxin 2; Syntaxin 3; Syntaxin 4; Synuclein alpha; VAMP-2; Vesicular Acetylcholine Transporter/VAChT; Vesicular GABA transporter/VGAT/VIAAT; Vesicular Glutamate Transporter 1, 2, 3/VGLUT; Vesicular monoamine transporter 1, 2 |
| Cholinergic |
Acetylcholine/ACh; Acetylcholinesterase; Choline Acetyltransferase/ChAT; Choline transporter; Vesicular Acetylcholine Transporter/VAChT |
| Dopaminergic |
Adrenaline; Dopamine; Dopamine Beta Hydroxylase/DBH; Dopamine Transporter/DAT; L-DOPA; Nitric Oxide-Dopamine; Norepinephrine; Norepinephrine Transporter/NET; Parkin; Tyrosine Hydroxylase/TH; TorsinA |
| Serotonergic |
DL-5-Hydroxytryptophan; Serotonin; Serotonin Transporter/SERT; Tryptophan Hydroxylase |
| GABAergic |
DARPP-32; GABA; GABA Transporters 1; GABA Transporters 2; GABA Transporters 3; Glutamate Decarboxylase/GAD; Vesicular GABA transporter/VGAT/VIAAT |
| Glutamatergic |
Glutamate; Glutamate Transporter; Glutamine; Glutamine Synthetase; Vesicular Glutamate Transporter 1; Vesicular Glutamate Transporter 2; Vesicular Glutamate Transporter 3 |
References
- ↑ Redwine, J. M.; Evans, C. F. (2002). "Markers of central nervous system glia and neurons in vivo during normal and pathological conditions". Current Topics in Microbiology and Immunology. 265: 119–40.
- ↑ Jefferis G, Livet J. Sparse and combinatorial neuron labelling. Curr Opin Neurobiol. 2012;22:101-10
- ↑ Swanger, SA; Bassell, GJ; Gross, C (2011). "High-resolution fluorescence in situ hybridization to detect mRNAs in neuronal compartments in vitro and in vivo". Methods Mol Biol. 714: 103–123.
- ↑ Graus, F.; Ferrer, I. (1990). "Analysis of a neuronal antigen (Hu) expression in the developing rat brain detected by autoantibodies from patients with paraneoplastic encephalomyelitis". Neuroscience Letters. 112 (1): 14–18. doi:10.1016/0304-3940(90)90314-Y.
- ↑ Wuenschell, C. W.; Fisher, R. S.; Kaufman, D. L.; Tobin (1986). "In situ hybridization to localize mRNA encoding the neurotransmitter synthetic enzyme glutamate decarboxylase in mouse cerebellum". Proceedings of the National Academy of Sciences of the United States of America. 83 (16): 6193–7. doi:10.1073/pnas.83.16.6193.
- ↑ Islam, M.R. (2007). In situ hybridization histochemistry: a novel method for neuronal tissues study. Bangl. J. Vet. Med. (2007). 5 (1 & 2): 111–114
- ↑ "Life and Discoveries of Camillo Golgi". nobelprize.org. Retrieved 2014-04-19.
- ↑ "Whonamedit - Franz Nissl". whonamedit.com. Retrieved 2014-04-19.
- ↑ Sherrington, C. S. (1935). "Santiago Ramon y Cajal. 1852–1934". Obituary Notices of Fellows of the Royal Society. 1 (4): 424. doi:10.1098/rsbm.1935.0007.
- ↑ "Albert Hewett Coons, June 28, 1912—September 30, 1978 | By Hugh O. McDevitt | Biographical Memoirs". books.nap.edu. Retrieved 2014-04-19.
- 1 2 3 4 Von Bohlen Und Halbach, O (2007). "Immunohistological markers for staging neurogenesis in adult hippocampus". Cell and tissue research. 329: 409–20. doi:10.1007/s00441-007-0432-4.
- ↑ Gall, J., & Pardue, M. (1969). Formation and detection of RNA-DNA hybrid molecules in cytological preparations. … of the National Academy of Sciences, (1), 378–383. Retrieved from http://www.pnas.org/content/63/2/378.short
- 1 2 Islam, M (2007). "In situ hybridization histochemistry: a novel method for neuronal tissues study". Bangladesh Journal of Veterinary Medicine. 5: 111–114. doi:10.3329/bjvm.v5i1.1327.
- ↑ Swanger, S. A.; Bassell, G. J.; Gross, C. (2011). "High-Resolution Fluorescence In Situ Hybridization to Detect mRNAs in Neuronal Compartments In Vitro and In Vivo". J. E. Gerst, Ed. 714: 103–123. doi:10.1007/978-1-61779-005-8.
- 1 2 3 4 5 Tanapat, Patima. "Neuronal Cell Markers". Materials and Methods. 3. doi:10.13070/mm.en.3.196.
- ↑ "In Situ Hybridization Histochemistry as a Supplement to Immunohistochemistry". In Situ Hybridization in Brain: 249–252. doi:10.1007/978-1-4615-9486-4_17. Retrieved 2014-04-19.
- 1 2 Purves, D., Augustine, G.J., Fitzpatrick, D., LaMantia, Anthony-Samuel, Hall, W.C., White, L.E. (2012). "22". Neuroscience (5 ed.). Sunderland, Massachusetts U.S.A: Sinauer Associates, INC.
- 1 2 Sharon A. Louis; Brent A. Reynolds (2013). "Neural Stem Cells" (PDF). STEMCELL Technologies.
- 1 2 Gage, F (2000). "Mammalian neural stem cells". Science. 287: 1433–1438. doi:10.1126/science.287.5457.1433.
- 1 2 "Neural Stem Cell Markers". R&D Systems.
- ↑ Leone, D. P.; Relvas, J. B.; Campos, L. S.; Hemmi, S.; Brakebusch, C.; Fässler, R.; Suter, U. (2005). "Regulation of neural progenitor proliferation and survival by beta1 integrins". Journal of Cell Science. 118 (12): 2589–99. doi:10.1242/jcs.02396.
- ↑ Wilson, P. G.; Stice, S. S. (2006). "Development and differentiation of neural rosettes derived from human embryonic stem cells". Stem Cell Reviews. 2 (1): 67–77. doi:10.1007/s12015-006-0011-1.
- ↑ Malatesta, P.; Appolloni, I.; Calzolari, F. (2008). "Radial glia and neural stem cells". Cell and Tissue Research. 331 (1): 165–78. doi:10.1007/s00441-007-0481-8.
- ↑ https://www.millipore.com/techpublications/tech2/nervous_system_markers
- ↑ "Neuro-Chrom™ Pan Neuronal Marker-Rabbit - Millipore". millipore.com. Retrieved 2014-04-19.
- ↑ Magavi, S.; Macklis, J. (2008). "Immunocytochemical analysis of neuronal differentiation". Neural Stem Cells. 198: 291–297.
- ↑ Lavezzi, A. M.; Corna, M. F.; Matturri, L. (2013). "Neuronal nuclear antigen (NeuN): a useful marker of neuronal immaturity in sudden unexplained perinatal death". Journal of the neurological sciences. 329: 45–50. doi:10.1016/j.jns.2013.03.012.
- ↑ Rusanescu, G.; Mao, J. (2016). "Peripheral nerve injury induces adult brain neurogenesis and remodeling". Journal of Cellular and Molecular Medicine. 20. doi:10.1111/jcmm.12965. PMID 27665307.
- 1 2 http://crm.nih.gov/stemcell_types/NSC/Differentiation_NSC.pdf
- ↑ "ChAT or Choline Acetyltransferase Antibody". neuromics.com. Retrieved 2014-04-19.
- ↑ Tian, C.; Liu, Q.; Ma, K.; Wang, Y.; Chen, Q.; Ambroz, R.; Zheng, J. C. (2013). "Characterization of induced neural progenitors from skin fibroblasts by a novel combination of defined factors". Scientific Reports. 3: 1345. doi:10.1038/srep01345.
- ↑ Perez, S. E.; Dar, S.; Ikonomovic, M. D.; Dekosky, S. T.; Elliott, J. (2008). "Cholinergic forebrain degeneration in the APPswe/PS1ΔE9 transgenic mouse". Neurobiology of Disease. 28 (1): 3–15. doi:10.1016/j.nbd.2007.06.015.