Pantothenate kinase
| Pantothenate kinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 2.7.1.33 | ||||||||
| CAS number | 9026-48-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Pantothenate kinase (EC 2.7.1.33, PanK; CoaA) is the first enzyme in the Coenzyme A biosynthetic pathway. It phosphorylates pantothenate (vitamin B5) to form 4'-phosphopantothenate at the expense of a molecule of adenosine triphosphate (ATP). It is the rate-limiting step in the biosynthesis of CoA.[1][2]

CoA is a necessary cofactor in all living organisms. It acts as the major acyl group carrier in many important cellular processes, such as the citric acid cycle (tricarboxylic acid cycle) and fatty acid metabolism. Consequently, pantothenate kinase is a key regulatory enzyme in the CoA biosynthetic pathway.[3]
Types
Three distinct types of PanK has been identified - PanK-I (found in bacteria), PanK-II (mainly found in eukaryotes, but also in the Staphylococci) and PanK-III, also known as CoaX (found in bacteria). Eukaryotic PanK-II enzymes often occur as different isoforms, such as PanK1, PanK2, PanK3 and PanK4. In humans, four PanK isoforms are expressed by three genes. PANK1 gene encodes the PanK1α and PanK1β forms, and PANK2 and PANK3 encode PanK2 and PanK3, respectively.[4]
Structure
PanK-II

PanK-II contains two protein domains, as illustrated in Figure 1. The A domain and A' domain each has a glycine-rich loop (sequence GXXXXGKS; P loop) that is characteristic of nucleotide-binding sites; this is where ATP is assumed to bind.[5] located between residues 95 and 102 on the A domain
The two ATP binding sites display cooperative behavior. The dimerization interface consists of two long helices, one from each monomer, that interact with each other. The C-terminal ends of the helices are held together by van der Waals interactions between valine and methionine residues of each monomer. The middle of the helices is attached by hydrogen bonds between asparagine residues. At the N-terminal end, each helix widens and forms a four-helix bundle with two shorter helices. This bundle consists of a hydrophobic core formed by non-polar residues that utilize van der Waals forces to further stabilize the dimer.[4]
In the active site, pantothenate is oriented by hydrogen bonds between pantothenate and the side chains of aspartate, tyrosine, histidine, tyrosine, and asparagine residues.[6] Asparagine, histidine, and arginine residues are involved in catalysis.
Human PanK-II isoforms PanK1α, PanK1β, PanK2, and PanK3 have a common, highly homologous catalytic core of approximately 355 residues.[4] PanK1α and PanK1β are both encoded by the PANK1 gene and have the same catalytic domain of 363 amino acids, encoded by exons 2 through 7. The PanK1α transcript starts with exon 1α that encodes a 184-residue regulatory domain at the N-terminus. This region allows for feedback inhibition by free CoA and acyl-CoA and regulation by acetyl-CoA and malonyl-CoA. On the other hand, the PanK1β transcript starts with exon 1β, which produces a 10-residue N-terminus that does not include a feedback regulatory domain.[7]
PanK-III

PanK-III also contains two protein domains, and the key catalytic residues of PanK-II are conserved. The monomer units of PanK-II and PanK-III are virtually identical, but they have distinctly different dimer assemblies. A study between the structures of Staphylococcus aureus type II and the Pseudomonas aeruginosa type III demonstrate that the PanK-II monomer has a loop region that is absent from the PanK-III monomer, and the PanK-III monomer has a loop region that is absent from the PanK-II monomer.[8] This minor variation has a crucial difference on the dimerization interface in which the helices of the PanK-II dimer coil around one another and the helices of the PanK-III dimer interact at a 70° angle (Figure 2).[9]
As a result of this difference in dimerization interface between PanK-II and PanK-III, the conformations of the substrate binding sites for ATP and pantothenate are also distinct.[10][11]
Catalytic Mechanism
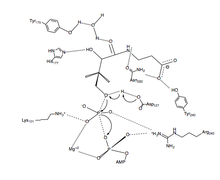
PanK-II
A proposed mechanism of the phosphoryl transfer reaction of PanK-II is a concerted mechanism with a dissociative transition state.
First, the ATP binds at the binding groove created by residues of the P loop and nearby residues. Here, the conserved lysine (Lys-101) is the key residue required for ATP binding.[12][13] Additionally, the side chains of residues Lys-101, Ser-102, Glu-199, and Arg-243 orient the nucleotide in the binding groove. The pantothenate is bound and oriented by forming hydrogen bond interactions with residues Asp-127, Tyr-240, Asn-282, Tyr-175, and His-177.[6] When both ATP and pantothenate are bound, Asp-127 deprotonates the C1 hydroxyl group of pantothenate. The oxygen from the pantothenate then attacks the γ-phosphate of the bound ATP. Here, charge stabilization of β- and γ-phosphate groups is achieved by Arg-243, Lys-101, and a coordinated Mg2+ ion.[14] In this concerted mechanism, the planar phosphorane of the γ-phosphate is transferred in-line to the attacking oxygen of pantothenate.[6] Finally, 4'-phosphopantothenate dissociates from PanK, followed by ADP.
Regulation of pantothenate kinase
PanK-II
The regulation of pantothenate kinase is essential to controlling the intracellular CoA concentration.[15] Pantothenate kinase is regulated via feedback inhibition by CoA and its thioesters (i.e., acetyl-CoA, malonyl-CoA).[16] CoA inhibits PanK activity by competitively binding to the ATP binding site and preventing ATP binding to Lys-101.[12][13]
Although CoA binds at the same site as ATP, they bind in distinct orientations, and their adenine moieties interact with the enzyme with nonoverlapping sets of residues. His-177, Phe-247, and Arg-106 are necessary for CoA recognition but not for ATP, and while Asn-43 and His-307 interact with the adenine base of ATP, His-177 and Phe-247 interact with the adenine base of CoA.[14] Both molecules use Lys-101 to neutralize the charge on their respective phosphodiesters.
Interestingly, nonesterified CoA has more potent inhibition than its thioesters. This phenomenon is best explained by the tight fit of the thiol group with the surrounding aromatic residues, Phe-244, Phe-259, Tyr-262, and Phe-252. Free CoA has an optimal fit, but when an acyl group is attached to CoA, the steric hindrance makes it difficult for the thioester to interact with Phe-252. Thus, the inhibition by thioesters is less effective than that by nonesterified CoA.[14]
PanK-III
The regulation outlined above corresponds to PanK-II. PanK-III is resistant to feedback inhibition.[8][10][11]
Genes
In humans:
The PANK2 gene encodes for PanK2, which regulates the formation of CoA in mitochondria, the cell’s energy-producing centers.[17] PANK2 mutation is the cause of Pantothenate kinase-associated neurodegeneration, formerly called Hallervorden-Spatz syndrome. This rare disease presents with profound dystonia, spasticity and is often fatal.
External links
- Pantothenate kinase at the US National Library of Medicine Medical Subject Headings (MeSH)
- EC 2.7.1.33
See also
References
- ↑ Robishaw, J. D.; Berkich, D.; Neely, J. R. (1982-09-25). "Rate-limiting step and control of coenzyme A synthesis in cardiac muscle". The Journal of Biological Chemistry. 257 (18): 10967–10972. ISSN 0021-9258. PMID 7107640.
- 1 2 Yang, Kun; Eyobo, Yvonne; Brand, Leisl A.; Martynowski, Dariusz; Tomchick, Diana; Strauss, Erick; Zhang, Hong (2006-08-01). "Crystal Structure of a Type III Pantothenate Kinase: Insight into the Mechanism of an Essential Coenzyme A Biosynthetic Enzyme Universally Distributed in Bacteria". Journal of Bacteriology. 188 (15): 5532–5540. doi:10.1128/JB.00469-06. ISSN 0021-9193. PMC 1540032
 . PMID 16855243.
. PMID 16855243. - ↑ Leonardi, Roberta; Zhang, Yong-Mei; Rock, Charles O.; Jackowski, Suzanne (2005-03-01). "Coenzyme A: Back in action". Progress in Lipid Research. 44 (2–3): 125–153. doi:10.1016/j.plipres.2005.04.001.
- 1 2 3 Hong, Bum Soo; Senisterra, Guillermo; Rabeh, Wael M.; Vedadi, Masoud; Leonardi, Roberta; Zhang, Yong-Mei; Rock, Charles O.; Jackowski, Suzanne; Park, Hee-Won (2007-09-21). "Crystal Structures of Human Pantothenate Kinases INSIGHTS INTO ALLOSTERIC REGULATION AND MUTATIONS LINKED TO A NEURODEGENERATION DISORDER". Journal of Biological Chemistry. 282 (38): 27984–27993. doi:10.1074/jbc.M701915200. ISSN 0021-9258. PMID 17631502.
- ↑ Saraste, Matti; Sibbald, Peter R.; Wittinghofer, Alfred (1990-11-01). "The P-loop — a common motif in ATP- and GTP-binding proteins". Trends in Biochemical Sciences. 15 (11): 430–434. doi:10.1016/0968-0004(90)90281-F.
- 1 2 3 Ivey, Robert A.; Zhang, Yong-Mei; Virga, Kristopher G.; Hevener, Kirk; Lee, Richard E.; Rock, Charles O.; Jackowski, Suzanne; Park, Hee-Won (2004-08-20). "The Structure of the Pantothenate Kinase·ADP·Pantothenate Ternary Complex Reveals the Relationship between the Binding Sites for Substrate, Allosteric Regulator, and Antimetabolites". Journal of Biological Chemistry. 279 (34): 35622–35629. doi:10.1074/jbc.M403152200. ISSN 0021-9258. PMID 15136582.
- ↑ Rock, Charles O; Karim, Mohammad A; Zhang, Yong-Mei; Jackowski, Suzanne (2002-05-29). "The murine pantothenate kinase (Pank1) gene encodes two differentially regulated pantothenate kinase isozymes". Gene. 291 (1–2): 35–43. doi:10.1016/S0378-1119(02)00564-4.
- 1 2 Yang, Kun; Strauss, Erick; Huerta, Carlos; Zhang, Hong. "Structural Basis for Substrate Binding and the Catalytic Mechanism of Type III Pantothenate Kinase †". Biochemistry. 47 (5): 1369–1380. doi:10.1021/bi7018578.
- ↑ Hong, Bum Soo; Yun, Mi Kyung; Zhang, Yong-Mei; Chohnan, Shigeru; Rock, Charles O.; White, Stephen W.; Jackowski, Suzanne; Park, Hee-Won; Leonardi, Roberta (2006-08-01). "Prokaryotic Type II and Type III Pantothenate Kinases: The Same Monomer Fold Creates Dimers with Distinct Catalytic Properties". Structure. 14 (8): 1251–1261. doi:10.1016/j.str.2006.06.008. ISSN 0969-2126. PMID 16905099.
- 1 2 Brand, Leisl A.; Strauss, Erick (2005-05-27). "Characterization of a New Pantothenate Kinase Isoform from Helicobacter pylori". Journal of Biological Chemistry. 280 (21): 20185–20188. doi:10.1074/jbc.C500044200. ISSN 0021-9258. PMID 15795230.
- 1 2 Choudhry, Anthony E.; Mandichak, Tracy L.; Broskey, John P.; Egolf, Richard W.; Kinsland, Cynthia; Begley, Tadhg P.; Seefeld, Mark A.; Ku, Thomas W.; Brown, James R. (2003-06-01). "Inhibitors of Pantothenate Kinase: Novel Antibiotics for Staphylococcal Infections". Antimicrobial Agents and Chemotherapy. 47 (6): 2051–2055. doi:10.1128/AAC.47.6.2051-2055.2003. ISSN 0066-4804. PMC 155856
 . PMID 12760898.
. PMID 12760898. - 1 2 Song, W. J.; Jackowski, S. (1994-10-28). "Kinetics and regulation of pantothenate kinase from Escherichia coli". The Journal of Biological Chemistry. 269 (43): 27051–27058. ISSN 0021-9258. PMID 7929447.
- 1 2 Song, W J; Jackowski, S (1992-10-01). "Cloning, sequencing, and expression of the pantothenate kinase (coaA) gene of Escherichia coli.". Journal of Bacteriology. 174 (20): 6411–6417. ISSN 0021-9193. PMC 207592
 . PMID 1328157.
. PMID 1328157. - 1 2 3 Yun, Mikyung; Park, Cheon-Gil; Kim, Ji-Yeon; Rock, Charles O.; Jackowski, Suzanne; Park, Hee-Won (2000-09-08). "Structural Basis for the Feedback Regulation of Escherichia coli Pantothenate Kinase by Coenzyme A". Journal of Biological Chemistry. 275 (36): 28093–28099. doi:10.1074/jbc.M003190200. ISSN 0021-9258. PMID 10862768.
- ↑ Jackowski, S.; Rock, C. O. (1981-12-01). "Regulation of coenzyme A biosynthesis". Journal of Bacteriology. 148 (3): 926–932. ISSN 0021-9193. PMC 216294
 . PMID 6796563.
. PMID 6796563. - ↑ Rock, Charles O.; Park, Hee-Won; Jackowski, Suzanne (2003-06-01). "Role of Feedback Regulation of Pantothenate Kinase (CoaA) in Control of Coenzyme A Levels in Escherichia coli". Journal of Bacteriology. 185 (11): 3410–3415. doi:10.1128/JB.185.11.3410-3415.2003. ISSN 0021-9193. PMC 155388
 . PMID 12754240.
. PMID 12754240. - ↑ "PANK2 gene". Genetics Home Reference. 2016-02-22. Retrieved 2016-02-29.