KDM2A
| KDM2A | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 |
|||||||||||||||
| |||||||||||||||
| Identifiers | |||||||||||||||
| Aliases | KDM2A, CXXC8, FBL11, FBL7, FBXL11, JHDM1A, LILINA, lysine demethylase 2A | ||||||||||||||
| External IDs | MGI: 1354736 HomoloGene: 56564 GeneCards: KDM2A | ||||||||||||||
| Targeted by Drug | |||||||||||||||
| daminozide[1] | |||||||||||||||
| RNA expression pattern | |||||||||||||||
 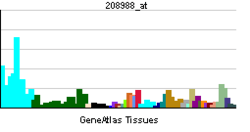  | |||||||||||||||
| More reference expression data | |||||||||||||||
| Orthologs | |||||||||||||||
| Species | Human | Mouse | |||||||||||||
| Entrez | |||||||||||||||
| Ensembl | |||||||||||||||
| UniProt | |||||||||||||||
| RefSeq (mRNA) | |||||||||||||||
| RefSeq (protein) | |||||||||||||||
| Location (UCSC) | Chr 11: 67.12 – 67.26 Mb | Chr 19: 4.31 – 4.4 Mb | |||||||||||||
| PubMed search | [2] | [3] | |||||||||||||
| Wikidata | |||||||||||||||
| View/Edit Human | View/Edit Mouse |
Lysine-specific demethylase 2A (KDM2A) also known as F-box and leucine-rich repeat protein 11 (FBXL11) is an enzyme that in humans is encoded by the KDM2A gene.[4][5][6]
Function
This gene encodes a member of the F-box protein family which is characterized by an approximately 40 amino acid motif, the F-box. The F-box proteins constitute one of the four subunits of ubiquitin protein ligase complex called SCFs (SKP1-cullin-F-box), which function in phosphorylation-dependent ubiquitination. The F-box proteins are divided into 3 classes: Fbws containing WD-40 domains, Fbls containing leucine-rich repeats, and Fbxs containing either different protein-protein interaction modules or no recognizable motifs. The protein encoded by this gene belongs to the Fbls class and, in addition to an F-box, contains at least 6 highly degenerated leucine-rich repeats.[6]
FBXL11/KDM2A is a histone H3 lysine 36 demethylase enzyme. The enzymatic activity of FBXL11/KDM2A relies on a conserved JmjC domain in the N-terminus of the protein that co-ordinates iron and alphaketoglutarate to catalyze demethylation via a hydroxylation based mechanism.[7] It has recently been demonstrated that a ZF-CxxC DNA binding domain within FBXL11/KDM2A has the capacity to interact with non-methylated DNA and this domain targets FBXL11/KDM2A to CpG island regions of the genome where it specifically removes histone H3 lysine 36 methylation.[8] This mechanism acts to create a chromatin environment at CpG islands that highlights these regulatory elements and differentiates them from non-regulatory regions in large complex mammalian genomes. In a study in mouse hepatocytes, this gene was shown to regulate hepatic gluconeogenesis.[9]
References
- ↑ "Drugs that physically interact with Lysine-specific demethylase 2A view/edit references on wikidata".
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (Jul 1999). "Prediction of the coding sequences of unidentified human genes. XIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro". DNA Res. 6 (1): 63–70. doi:10.1093/dnares/6.1.63. PMID 10231032.
- ↑ Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW (Dec 1999). "A family of mammalian F-box proteins". Curr Biol. 9 (20): 1180–2. doi:10.1016/S0960-9822(00)80021-4. PMID 10531037.
- 1 2 "Entrez Gene: FBXL11 F-box and leucine-rich repeat protein 11".
- ↑ Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (February 2006). "Histone demethylation by a family of JmjC domain-containing proteins". Nature. 439 (7078): 811–6. doi:10.1038/nature04433. PMID 16362057.
- ↑ Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ (Apr 2010). "CpG islands recruit a histone H3 lysine 36 demethylase". Molecular Cell. 38 (2): 179–90. doi:10.1016/j.molcel.2010.04.009. PMC 3098377
 . PMID 20417597.
. PMID 20417597. - ↑ Pan D, Mao C, Zou T, Yao AY, Cooper MP, Boyartchuk V, Wang YX (2012). "The Histone Demethylase Jhdm1a Regulates Hepatic Gluconeogenesis". PLoS Genetics. 8 (6): e1002761. doi:10.1371/journal.pgen.1002761. PMC 3375226
 . PMID 22719268.
. PMID 22719268.
Further reading
- Nakajima D, Okazaki N, Yamakawa H, Kikuno R, Ohara O, Nagase T (2003). "Construction of expression-ready cDNA clones for KIAA genes: manual curation of 330 KIAA cDNA clones.". DNA Res. 9 (3): 99–106. doi:10.1093/dnares/9.3.99. PMID 12168954.
- Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M (1999). "Identification of a family of human F-box proteins.". Curr. Biol. 9 (20): 1177–9. doi:10.1016/S0960-9822(00)80020-2. PMID 10531035.
- Ilyin GP, Rialland M, Pigeon C, Guguen-Guillouzo C (2001). "cDNA cloning and expression analysis of new members of the mammalian F-box protein family.". Genomics. 67 (1): 40–7. doi:10.1006/geno.2000.6211. PMID 10945468.
- Hattori A, Okumura K, Nagase T, Kikuno R, Hirosawa M, Ohara O (2001). "Characterization of long cDNA clones from human adult spleen.". DNA Res. 7 (6): 357–66. doi:10.1093/dnares/7.6.357. PMID 11214971.
- Watanabe N, Arai H, Nishihara Y, Taniguchi M, Watanabe N, Hunter T, Osada H (2004). "M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP.". Proc. Natl. Acad. Sci. U.S.A. 101 (13): 4419–24. doi:10.1073/pnas.0307700101. PMC 384762
 . PMID 15070733.
. PMID 15070733. - Colland F, Jacq X, Trouplin V, Mougin C, Groizeleau C, Hamburger A, Meil A, Wojcik J, Legrain P, Gauthier JM (2004). "Functional proteomics mapping of a human signaling pathway.". Genome Res. 14 (7): 1324–32. doi:10.1101/gr.2334104. PMC 442148
 . PMID 15231748.
. PMID 15231748. - Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M (2005). "Nucleolar proteome dynamics.". Nature. 433 (7021): 77–83. doi:10.1038/nature03207. PMID 15635413.
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006). "Histone demethylation by a family of JmjC domain-containing proteins.". Nature. 439 (7078): 811–6. doi:10.1038/nature04433. PMID 16362057.
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks.". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983.