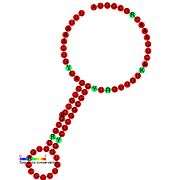
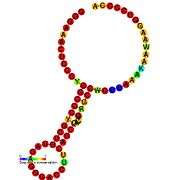
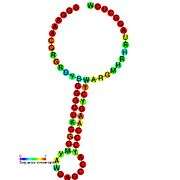
Ribosomal frameshift
A "ribosomal frameshift" allows alternative translation of an mRNA sequence by changing the open reading frame. This technique is commonly found in viruses, as it allows the virus to encode multiple types of proteins from the same mRNA. Frameshifting allows viruses to create many protein structures from a relatively small genome. While the ribosome is translating mRNA it may shift forward or backward, depending on the virus. Because three consecutive mRNA nucleotides make a codon that's translated into one amino acid, the frameshift makes every subsequent amino acid different because it translates a different set of codons in the new reading frame, resulting in a different protein product.
There are two kinds of frameshifting: pseudoknot structure and slippery sequence (usually UUUAAAC).[1] A pseudoknot is an RNA secondary structure generated by intermolecular interactions of mRNA that cause it to bend and fold. The ribosome bumps into the knotted mRNA while it is translating the mRNA and shifts back (-1) one nucleotide and continues translating in a new reading frame. This frameshift occurs about 10% of the time, allowing differential protein expression. This form of frameshifting is used by HIV-1, among others.[2] A slippery sequence is a sequence of codons for rare tRNA, meaning that the ribosome has to pause in order to wait for the correct tRNA to bring the appropriate amino acid. However, this long pause is often accompanied by the ribosome advancing forward one nucleotide (+1). This form of frameshifting is utilized by influenza A, among others.[3]
Other viruses that use ribosomal frameshifting for alternative protein expression include the barley yellow dwarf virus,[4] potato leafroll virus,[5] simian retrovirus-1, and coronaviruses.[6]
See also
- Translational frameshift
- HIV Ribosomal frameshift signal
- Coronavirus frameshifting stimulation element
- Ribosomal pause
References
- ↑ Meulenberg JJ, Hulst MM, de Meijer EJ, et al. (January 1993). "Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV". Virology. 192 (1): 62–72. doi:10.1006/viro.1993.1008. PMID 8517032.
- ↑ "Ribosomal Frameshifting". Viralzone.expasy.org. Swiss Institute of Bioinformatics. Retrieved 4 December 2014.
- ↑ "Ribosomal Frameshifting". Viralzone.expasy.org. Swiss Institute of Bioinformatics. Retrieved 4 December 2014.
- ↑ Vincent JR, Lister RM, Larkins BA (October 1991). "Nucleotide sequence analysis and genomic organization of the NY-RPV isolate of barley yellow dwarf virus". J. Gen. Virol. 72 (10): 2347–55. doi:10.1099/0022-1317-72-10-2347. PMID 1840612.
- ↑ Kujawa AB, Drugeon G, Hulanicka D, Haenni AL (May 1993). "Structural requirements for efficient translational frameshifting in the synthesis of the putative viral RNA-dependent RNA polymerase of potato leafroll virus". Nucleic Acids Res. 21 (9): 2165–71. doi:10.1093/nar/21.9.2165. PMC 309480
 . PMID 8502558.
. PMID 8502558. - ↑ Sung D, Kang H (March 1998). "Mutational analysis of the RNA pseudoknot involved in efficient ribosomal frameshifting in simian retrovirus-1". Nucleic Acids Res. 26 (6): 1369–72. doi:10.1093/nar/26.6.1369. PMC 147434
 . PMID 9490779.
. PMID 9490779.