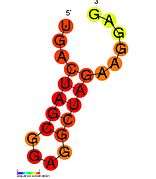
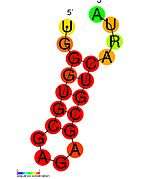
Retroviral Psi packaging element
| Human immunodeficiency virus type 1 dimerisation initiation site | |
|---|---|
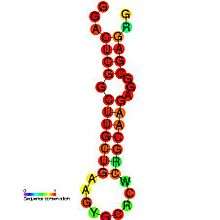 | |
| Predicted secondary structure and sequence conservation of HIV-1_DIS | |
| Identifiers | |
| Symbol | HIV-1_DIS |
| Alt. Symbols | Retroviral_psi |
| Rfam | RF00175 |
| Other data | |
| RNA type | Cis-regp |
| Domain(s) | Viruses |
| SO | 0000233 |
Retroviral Psi packaging element is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV)[1] and Simian immunodeficiency virus (SIV).[2] It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication.[3][4][5][6] The final virion contains a dimer of two identical unspliced copies of the viral genome.

In HIV, the Psi element is around 80–150 nucleotides in length, and located at the 5' end of the genome just upstream of the gag initiation codon.[8] It has a known secondary structure composed of four hairpins called SL1 to SL4 (SL is for Stem-loop) which are connected by relatively short linkers. All four stem loops are important for genome packaging and each of the stem loops SL1,[8] SL2,[9] SL3 [10][11] and SL4 [12] has been independently expressed and structurally characterised.
Stem loop 1 (SL1) (also referred to as HIV-1_DIS) consists of a conserved hairpin structure with a palindromic loop sequence which was predicted and confirmed by mutagenesis.[13] This palindromic loop is known as the primary dimer initiation site (DIS) as it is believed to promote dimerization of the viral genome through formation of a "kissing dimer" intermediate.[14] The Rfam structure shown is based on a covariation model.
It has been shown that SL1 may provide a secondary binding site for the viral Rev protein.[15] The Rev protein is an essential HIV regulatory protein which increases the stability and transport of the unspliced viral RNA.[16]
Stem-loop 2 (SL2) (also referred to as HIV-1 SD) consists of a highly conserved 19 nt stem-loop which has been shown by mutagenis to modulate the splicing efficiency of HIV-1 mRNAs.[17]
Stem-loop 3 (SL3) consists of a highly conserved 14 nt stem-loop which was predicted and confirmed by mutagenesis and mass spectrometric detection (MS3D). HIV-1 SL3 is sufficient by itself to induce heterologous RNA into virus-like particles but its deletion does not completely eliminate encapsidation.[17]
Stem-loop 4 (SL4) consists of a highly conserved 14 nt stem-loop that is located just downstream of the gag start codon. The structure was confirmed by mutagenesis and has an NMR and mass spectrometric detection (MS3D).[17] It also may have coding and non-coding roles.
References
- ↑ Lever A, Gottlinger H, Haseltine W, Sodroski J (1989). "Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions". J Virol. 63 (9): 4085–7. PMC 251012
 . PMID 2760989.
. PMID 2760989. - ↑ Strappe PM, Greatorex J, Thomas J, Biswas P, McCann E, Lever AM (2003). "The packaging signal of simian immunodeficiency virus is upstream of the major splice donor at a distance from the RNA cap site similar to that of human immunodeficiency virus types 1 and 2". J Gen Virol. 84 (9): 2423–30. doi:10.1099/vir.0.19185-0.
- ↑ McBride MS, Panganiban AT (1997). "Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo". J. Virol. 71 (3): 2050–8. PMC 191293
 . PMID 9032337.
. PMID 9032337. - ↑ McBride MS, Schwartz MD, Panganiban AT (1997). "Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation". J. Virol. 71 (6): 4544–54. PMC 191676
 . PMID 9151848.
. PMID 9151848. - ↑ McBride MS, Panganiban AT (1996). "The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures". J. Virol. 70 (5): 2963–73. PMC 190155
 . PMID 8627772.
. PMID 8627772. - ↑ Lever AM (2007). "HIV-1 RNA packaging". Adv. Pharmacol. Advances in Pharmacology. 55: 1–32. doi:10.1016/S1054-3589(07)55001-5. ISBN 978-0-12-373610-9. PMID 17586311.
- ↑ Baba S, Takahashi K, Noguchi S, Takaku H, Koyanagi Y, Yamamoto N, Kawai G (2005). "Solution RNA structures of the HIV-1 dimerization initiation site in the kissing-loop and extended-duplex dimers.". J Biochem. 138 (5): 583–92. doi:10.1093/jb/mvi158. PMID 16272570.
- 1 2 Lawrence DC, Stover CC, Noznitsky J, Wu Z, Summers MF (2003). "Structure of the intact stem and bulge of HIV-1 Psi-RNA stem-loop SL1". J. Mol. Biol. 326 (2): 529–42. doi:10.1016/S0022-2836(02)01305-0. PMID 12559920.
- ↑ Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF (2000). "NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition". J. Mol. Biol. 301 (2): 491–511. doi:10.1006/jmbi.2000.3979. PMID 10926523.
- ↑ Zeffman A, Hassard S, Varani G, Lever A (2000). "The major HIV-1 packaging signal is an extended bulged stem loop whose structure is altered on interaction with the Gag polyprotein". J Mol Biol. 297 (4): 877–93. doi:10.1006/jmbi.2000.3611. PMID 10736224.
- ↑ Pappalardo L, Kerwood DJ, Pelczer I, Borer PN (1998). "Three-dimensional folding of an RNA hairpin required for packaging HIV-1". J. Mol. Biol. 282 (4): 801–18. doi:10.1006/jmbi.1998.2046. PMID 9743628.
- ↑ Kerwood DJ, Cavaluzzi MJ, Borer PN (2001). "Structure of SL4 RNA from the HIV-1 packaging signal". Biochemistry. 40 (48): 14518–29. doi:10.1021/bi0111909. PMID 11724565.
- ↑ Berkhout, B; Van Wamel, JL (1996). "Role of the DIS hairpin in replication of human immunodeficiency virus type 1". Journal of Virology. 70 (10): 6723–32. PMC 190715
 . PMID 8794309.
. PMID 8794309. - ↑ Skripkin, E.; Paillart, J. C.; Marquet, R.; Ehresmann, B.; Ehresmann, C. (1994). "Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro". Proceedings of the National Academy of Sciences of the United States of America. 91 (11): 4945–4949. doi:10.1073/pnas.91.11.4945. PMC 43906
 . PMID 8197162.
. PMID 8197162. - ↑ Gallego J, Greatorex J, Zhang H, et al. (2003). "Rev binds specifically to a purine loop in the SL1 region of the HIV-1 leader RNA". J. Biol. Chem. 278 (41): 40385–91. doi:10.1074/jbc.M301041200. PMID 12851400.
- ↑ Felber, B. K.; Hadzopoulou-Cladaras, M.; Cladaras, C.; Copeland, T.; Pavlakis, G. N. (1989). "Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA". Proceedings of the National Academy of Sciences of the United States of America. 86 (5): 1495–1499. doi:10.1073/pnas.86.5.1495. PMC 286723
 . PMID 2784208.
. PMID 2784208. - 1 2 3 Abbink, T. E. M.; Berkhout, B. (2007). "RNA Structure Modulates Splicing Efficiency at the Human Immunodeficiency Virus Type 1 Major Splice Donor". Journal of Virology. 82 (6): 3090–3098. doi:10.1128/JVI.01479-07. PMC 2258995
 . PMID 18160437.
. PMID 18160437.
External links
- Page for HIV-1_DIS at Rfam
- Page for HIV-1_SD at Rfam
- Page for HIV-1 SL3 at Rfam
- Page for HIV-1 SL4 at Rfam