Lysine 2,3-aminomutase
| Lysine 2,3-aminomutase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 5.4.3.2 | ||||||||
| CAS number | 9075-20-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Lysine 2,3-aminomutase (KAM or LAM) (EC 5.4.3.2) is a radical SAM enzyme that facilitates the conversion of the amino acid lysine to beta-lysine.[1][2] [3][4] It accomplishes this interconversion using three cofactors and a 5'-deoxyadenosyl radical formed in a S-Adenosyl methionine (SAM) activated radical reaction pathway.[1] The generalized reaction is shown below:
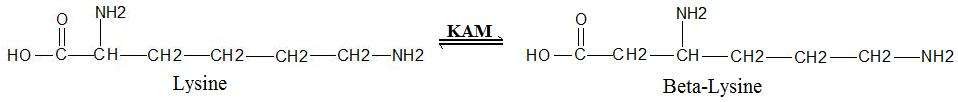
Structure
Shown on the right is the three-dimensional structure of the Lysine 2,3-aminomutase protein. The structure was determined by X-ray crystallography to 2.1 Angstrom resolution and was seen to crystallize as a homotetramer.[2] KAM was first purified and characterized in Clostridium subterminale for studies of Lysine metabolism.
Cofactors
Three key cofactors are required for the reaction catalyzed by the lysine 2,3-aminomutase enzyme. They are:
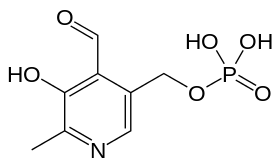
- Pyridoxal phosphate (PLP): Responsible for binding of the amino acid during reaction. The pi-system of this molecule facilitates radical delocalization during formation of an aziridinyl radical. The structure is given below:
- Zinc metal: Required for coordination between the dimers in the protein.
- Iron-sulfur cluster: A 4 iron-4 sulfur cluster is required for formation of a 5'-deoxyadenosyl radical. This radical then acts as the "stable" radical carrier in the reaction mechanism which transfers the radical to the amino acid.
Reaction Mechanism
The generalized reaction takes place in 5 steps:
- Radical Formation: A "stable" radical is formed through a radical SAM mechanism in which a S-adenosyl methionine forms a 5'-deoxyadenosyl radical.
- Enzyme Binding: Lysine 2,3-aminomutase binds to pyridoxal phosphate (PLP).
- Amino Acid Binding: The amino acid (Lysine or Beta-Lysine depending on forward or reverse reactions) binds to pyridoxal phosphate.
- Radical Transfer: The 5'-deoxyadenosyl radical is transferred to the amino acid and an aziridinyl radical is formed. In this configuration, the radical is stabilized by the pi-system of pyridoxal phosphate.
- Amino Acid Conversion: In the final step, the new amino acid is formed and the radical is returned to its more stable state on the 5'-deoxyadenosyl.
The reaction mechanism described above is shown below:

References
- ↑ Frey, P.A. "Lysine 2,3-aminomutase: is adenosylmethionine a poor man's adenosylcobalamin. FASEB Jour.; 1993; Vol. 7, 662-670.
- ↑ Lepore, B.; Ruzicka, F.J.; Frey, P.A.; Ringe, D. "The x-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale. PNAS; 2005; 102, 13819-13824.
- ↑ Aberhart, D.J.; Lim, H.-J.; Weiller, B.H. (1981). "Stereochemistry of lysine 2,3-aminomutase". J. Am. Chem. Soc. 103: 6750–6752. doi:10.1021/ja00412a040.
- ↑ Zappia, V.; Barker, H.A. (1970). "Studies on lysine-2,3-aminomutase. Subunit structure and sulfhydryl groups". Biochim. Biophys. Acta. 207: 505–513. doi:10.1016/s0005-2795(70)80013-7. PMID 5452674.
External links
- Lysine 2,3-aminomutase at the US National Library of Medicine Medical Subject Headings (MeSH)