Boom method
Boom method (Boom nucleic acid extraction method) is a Solid phase extraction method for isolating nucleic acid from a biological sample. This method is characterized by "Absorbing the nucleic acids (NA) to the Silica beads".
Overview
Boom method (Boom nucleic acid extraction method) [1] [2] [3][4][5] [6] [7] [8] is a solid phase extraction method for "isolating nucleic acid (NA) [notes 1][9] from Biological samples. Essential of this method is the use of Silica beads, capable of binding the NA in the presence of a chaotropic substance according to the effect. This method is one of the most widespread [6][7] methods for isolating nucleic acids from biological samples and is known as a simple, rapid, and reliable [2] method for the small-scale purification of NA from Biological sample.
This method is said to have been developed and invented by Willem R. Boom et al. around 1990 .[notes 2] However, the aforementioned chaotropic effect itself was already known and already reported by Vogelstein and Gillespie [10][notes 3] before the development of BOOM method. So the contribution of Boom et al. may be the optimization of the method to complex starting materials.[1] , such as body fluids and other biological starting materials, and provides a short step procedure according to the Boom et. al US5234809.[1] And, little later from the Boom et.al [1] is filed, similar application such as ,[11] [12] have also been filed by other parties
In narrow sense, the word "Silica" meant SiO2 crystals however, other forms of Silica particles are available. Especially amorphous silicon oxide and glass powder, alkylsilica, aluminum silicate (zeolite), or, activated silica with -NH2, are suitable as nucleic acid binding solid phase material according to this method. Today, the embodyments of Boom method, characterized by "utilizing the magnetic beads (silica beads are magnetic beads)" is widely used. In such method, silica beads are captured by magnetic beads collector, such as Tajima Pipette [13] [14] [15] , Pick pen(R),[3][4] Quad Pole collector,[16] ..., and so on.
Brief procedure and Basic Principle are described below.
Brief procedure

The process for isolating nucleic acid from Starting material of Boom method are essentially consist of following 4 steps [1] [2] [3] [4] (See Fig.1.).
(a)Lysing and/or Homogenizing the Starting material.
Lysate of Starting material is obtained by for example a detergent in the presence of protein degrading enzymes.(b)Mixing chaotropic substance and Silica beads into the Starting material.
Mixing the Starting material', a chaotropic substance to bind the NA to Silica beads, Lysate of Starting material of (a) is mixed with sufficiently large amounts of chaotropic substance. According to the chaotropic effect, releasing-NA will be bound to the silica beads almost instantaneously. In this way, silica-nucleic acid complexes are formed. The reasons "why NA and Silica form bond" are to be described in following section (Basic Principals).(c)Washing Silica beads
In this step, Silica beads of (b) are washed several times to remove contaminants. Process of Washing of the silica-nucleic acid complexes (silica beads) typically consists of following steps,
- Collecting Silica beads from the liquid by for example Tajima pipette (see Fig. 1,2) or by for example Pellet-down (by rapid sedimentation (centrifugation) and disposal of the supernatant (e.g., bysuction))
- Redispersioning Silica beads into the chaotropic salt-containing washing buffer using, e. g., a vortex mixer.
- Collecting redispersed Silica beads from above mentioned washing buffer again.
- Further washed successively with an alcohol-water solution [notes 4] and with acetone.
- Drying beads are preferably be done.
(d)Separating the bonded nucleic acids
Separating the bonded nucleic acids from the Silica beads. Pure NA are eluted into buffer by decreasing the concentration of chaotropic substance. NA presented in the washed (and preferably dried) silica-nucleic acid complexes is eluted into elution buffer such as TE buffer, aqua bidest, ... , and so on. The selection of the elution buffer is co-determined by the contemplated use of the isolated NA.
In this way, pure NA are isolated from the Starling material.
By the alteration of the experimental condition, especially by alteration of the composition of reagents (chaotropic substance, wash buffer, and so on) we can realize more specific isolation. For example, some composition of reagents are suitable for obtaining long ds DNA, some composition of reagents are suitable for short ss RNA,..., and so on.
Starting material are for example, whole blood, blood serum, buffy coat, urine, feces, liquor cerebrospinal, sperm, saliva, tissues, cell cultures, foods products, vaccines. and..., so various Starting biological material are available.
Of cause optimization of procedure according to the Starting material, species of desired nucleic acid (DNA/RNA, Linear/ circular, ds/ss, long/short) are required.
Today, the assay characterized by using Silica coated magnetic beads seems the mainstream. So, in this article Silica beads are intended to mean the Silica coated magnetic beads unless special annotation.
Magnetic beads
The silica coated beads [17] [18] [19] , coated various magnetic particles (magnetic carrier) with silica are often used. Maghemite particle (γ-Fe2O3) and magnetite particle (Fe3O4), as well as an intermediate iron oxide particle thereof, are most suitable as magnetic carrier.
Generally, the quality of the magnetic beads are characterized by following parameters [17]
- saturation magnetization (~10-80 A m2/kg (emu/g):Superparamagnetic),[17]
- coercive force (~ 0.80-15.92 kA/m),[17]
- size diameter (~ 0.1-0.5 μm),[17]
- mass of each particle (~ 2.7 ng),[17][notes 5]
- ease of collection (to be mentioned later),[17]
- capture ability (to be mentioned later),[17]
- Sedimentation rate (~4% in 30 min),[20]
- Area ratio(> 100 m2/g),
- Effective density(~ 2.5 g/cm3), and
- Particle counts (~ 1 x 108 particles/mg).[20]
Here, "ease of collection" are defined and compared by
"magnetic beads are collected by not less than X wt % (~90wt %) within T seconds(~ 3 seconds) in the presence of a magnetic field of Y gauss (~3000 gauss) when it is dispersed in an amount of at least Z mg (~20 mg) in W mL (~1 mL) of an aqueous solution of a sample containing a biological substance"
and, capture ability are defined and compared by
"binding with at least A μg (~0.4μg) of the biological substance per B mg (~1 mg) thereof when it is dispersed in an amount of at least Z mg (~20 mg) in W mL (~1 mL) of an aqueous solution of a sample containing a biological substance".
Basic Principles
The principal of this method [1] [2] [3] [4] [13] is based on the nucleic acid-binding properties of silica particles or diatoms in the presence of this Chaotropic agent, which are according to the Chaotropic effect. In this section, we will explain the mechanism of the "nucleic acid-binding properties of silica under the Chaotropic agent" in view of Chaotropic effect.
Put simply, Chaotropic effect is where a Chaotropic anion in aqueous solution disturbing the structure of water, and weakens the hydrophobic interaction [21] .[22]
In the broad sense, Chaotropic agent stands for any substance capable of altering the secondary, tertiary and/or quaternary structure of proteins and nucleic acids, but leaving at least the primary structure intact.[23]
Aqueous solution of Chaotropic salt is a Chaotropic agent. Chaotropic anion increase the entropy of the system by interfering with inter molecular interactions mediated by non-covalent forces such as hydrogen bonds, van der Waals forces, and hydrophobic effects. Examples thereof are aqueous solution of: Thiocyanate ion, Iodine ion, Perchlorate ion, Nitrate ion, Bromine ion, Chlorine ion, Acetate ion, Fluorine ion, and Sulfate ion, or mutual combinations therewith. According to the original method of Boom method, the chaotropic guanidinium salt employed IS preferably guanidinium thiocyanate (GuSCN).
According to the Chaotropic effect, in the presence of the chaotropic agent, hydration water of NA are taken from the Phosphodiester bond of the Phosphate group of the skeleton of a NA thereby, the phosphate group becomes "exposed" therefore, hydrophobic interaction between silica and exposed phosphate group are formed.
Automated Instruments
Tajima Pipette
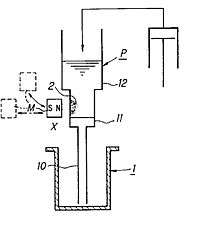
Nucleic acid extraction apparatus based on Tajima Pipette[13] [14] (see Fig.2) are one of the most widespread instruments to perform the Boom method.[24]
Tajima Pipette was invented by Hideji Tajima (jp),[13] he is founder president of Precision System Sciences (PSS) [24] Inc., a Japanese manufacturer of precision and measuring instruments. Tajima Pipette is a Core Technology of PSS Inc.[24] PSS Inc. provides OEM product based on this technology (for example MagNA Pure(R) ) for several leading reagent manufacturers such as Hoffmann-La Roche, Life Technologies, ... and so on. Little later from the Tajima et al.[13] is filed, similar application such as [15] have also been filed by other parties.
Tajima Pipette performs magnetic particle control method and procedure, which can separates magnetic particles combined with a target substance from the liquid by magnetic force and, which can suspends them in a liquid.
Configurations
Tajima pipette itself is an apparatus comprising following members (see Fig.2).[13]
- pipette tip configured to be able to access and aspirate/discharge liquid from/into each of vessels, having
- a front end portion,
- a reservoir portion,
- a liquid passage
- connecting the front end portion and the reservoir portion,
- a separation region
- in the liquid passage subjected to an action of a magnetic field, and
- a mechanism
- for applying a negative or positive pressure to the interior of the pipette portion to draw or discharge a magnetic substance suspended liquid into or from the pipette portion
- magnetic field Source
- arranged on the outside of and adjacent to pipette tip; and
- magnetic field source driving device
- for driving the magnetic field source to apply or remove a magnetic field to or from the separation region from outside the liquid passage. When magnet is brought close to the pipette tip , magnetic field areapplied, and retract away from the pipette tip, that magnetic field are removed.
Nucleic acid extraction apparatus which incorporates Tajima pipette are typically consists of [13]
- Above mentioned Tajima pipette,
- Plurality of tubes.
- Plurality of tube holder for above mentioned tubes,
- Transport mean
- to transport Tajima pipette among that plurality of tubes (tubes are supported by tube holder), and
- Control device
- for controlling abovementioned devices.
Motions
(a)Capturing of the magnetic beads.
During this suction process,
when the magnetic field are applied to the separation region of piper tip, from outside of pipette tip, by the magnet arranged on the outside of the pipette tip,
as liquid containing magnetic beads passes through a separation region of the pipette tip,
the magnetic particles are attracted to and arrested to the inner wall of tile separation region of pipette tip.
Subsequently, when that solution are discharged under the conditions of has been kept the magnetic field, magnetic particles only are left in the inside of pipette tip. In this way magnetic particles are separated from liquid.
In accordance with Tajima,[13] the preferable suction height of the mixture liquid is such that
- the bottom level of the liquid is higher than the lower end of the separation region of the liquid passage (That means bottom level of the liquid is higher than the lower end of the magnet.),
- when all the mixture liquid is drawn up,
- so as to ensure that the aspirated magnetic particles can be completely arrested.
At this time, because the magnetic particles are wet, they stay attached to the inner surface of the separation region of the liquid passage of the pipette tip. If the pipette tip P is moved or transported, the magnetic particles will not come off easily.
(b)Re-suspension of the captured magnetic beads.
After the magnetic particles are arrested by above mentioned manner (a),
- so the mixture liquid removed of the magnetic particles is discharged into the liquid accommodating portion (Vessel) and drained out, with only the magnetic particles remaining in the pipette tip,
we can do the re-suspension process.
Re-suspension of the captured magnetic beads are in detail, consists of the following steps. Of cause, we consider that, the state in which that magnetic material has been captured by above mention way.
- Aspirate liquid such as washing buffer into the tip
- Quit the application of a magnetic field
- By "Quit the application of a magnetic field" the magnetic particles are suspended in the liquid.
- Discharging the liquid (such as washing buffer) from pipette tip to vessel (in the condition of magnetic force generated by the magnet body is cut off.).
Operations
An example of the operations of the Nucleic acid extnlction apparatus which incorporates Tajima pipette are typically as shown in Fig. 1.
Other methods
Examplcs of other type of method of the magnetic particle capturing device are as follows.
Notes
- ↑ In this article, nucleic acid (NA) is meant both DNA and RNA, both in any possible configuration (double-stranded (ds) /single-stranded (ss) /as a combination thereof, Long/Short, Circular/Liner,...).
- ↑ Estimated from priority date of Boom, et. al; US5234809 , EP0389063 and their family patents.
- ↑ The following articles was cited in the "Boom, et. al; US5234809 , EP0389063 and their family patents".
- B Vogelstein and D Gillespie ; "Preparative and analytical purification of DNA from agarose" PNAS 1979 vol. 76 no. 2 pp.615-619
- ↑ According original method by Boom, most preferably about 70% ethanol to restrict losses in yield
- ↑ Order estimation procedures for estimating are as follows.
- Radius of each particle (r) is about
- 0.5 μm .
- So, volume of each particle (V) is about 5.2*10−13 cm3 by
- .
- When we supposed that magnatic particle are Fe3O4 then, the density (D) 5.17 g/cm3.
- So weight (w) of each particle is about
- w=VD=2.7 pg
References
- 1 2 3 4 5 6 Boom, et. al; US5234809 , EP0389063 and their family patents.
- 1 2 3 4 R Boom, C J Sol, M M Salimans, C L Jansen, P M Wertheim-van Dillen and J van der Noordaa;"Rapid and simple method for purification of nucleic acids."J. Clin. Microbiol. March 1990 vol. 28 no. 3 495-503
- 1 2 3 4 5 Matti Korpela; US6468810
- 1 2 3 4 5 Technical Notes by Bio-Nobile brand
- ↑ By John Brunstein;"Sample extraction methods: how we obtain DNA and RNA"
- 1 2 https://www.hanc.info/labs/labresources/procedures/ACTGIMPAACT%20Lab%20Manual/Standard%20Roche%20Monitor%20Test,%20Boom%20Extraction.pdf[]
- 1 2 http://www.siemens.com/about/pool/de/unser_geschaeft/healthcare/diagnostics/perspectives-assay-performance.pdf
- ↑ How do SPRI beads work?
- ↑ DNA Extraction Methods For Large Blood Volumes
- ↑ B Vogelstein and D Gillespie;"Preparative and analytical purification of DNA from agarose" PNAS 1979 vol. 76 no. 2 pp.615-619
- ↑ , US5973138 US5342931
- ↑ US5705628
- 1 2 3 4 5 6 7 8 9 10 Hideji Tajima;US5702950 , US6331277 ,
US 2001/0007770 A1 and their family patents.
See also .
Apparatus and methods on this patent are mainly intended to the immune system. However, alternative embodiment for nucleic acids extraction assay are also mentioned in this patent and procedure of Fig.1 is generally similar to Boom method. - 1 2 Hideji Tajima;US6509193
Apparatus and methods on this patent are mainly intended to the immune system. However, alternative embodiment for nucleic acids extraction assay are also mentioned in this patent and procedure of Fig.1 is generally similar to Boom method. - 1 2 Urban Schmitt et.al;US6187270
- 1 2 Paul A. Liberti et.al, US5466574
- 1 2 3 4 5 6 7 8 US20040126902
- ↑ US7119194
- ↑ US6368800
- 1 2 3 Magnetic Particles Archived December 3, 2013, at the Wayback Machine.(Written in Japanese)
- ↑ Web page of bio.davidson.edu
- ↑ maxXbond: first regeneration system for DNA binding silica matrices KH Esser, WH Marx, T Lisowsky – Nature Methods| Application Notes, 2006 see also,
- ↑ http://www.merriam-webster.com/medical/chaotropic
- 1 2 3 See the web site of Precision System Sciences (PSS) Inc.(Written in Japanese). Web site of their U.S branch are
- ↑ US7741129
- ↑ US7384559
- ↑ US6764859
See also
- Nucleic acid methods
- DNA extraction
- RNA extraction
- Ethanol precipitation
- Phenol-chloroform extraction
- Chaotropic agent
- DNA separation by silica adsorption
- minicolumn purification