Xanthomonas
| Xanthomonas | |
|---|---|
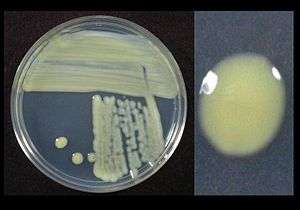 | |
| Xanthomonas translucens growing on sucrose peptone agar showing yellow pigment | |
| Scientific classification | |
| Kingdom: | Bacteria |
| Phylum: | Proteobacteria |
| Class: | Gammaproteobacteria |
| Order: | Xanthomonadales |
| Family: | Xanthomonadaceae |
| Genus: | Xanthomonas Dowson 1939 |
| Species | |
|
X. albilineans | |
Xanthomonas is a genus of Proteobacteria, many of which cause plant diseases.
Taxonomy
The Xanthomonas genus has been subject of numerous taxonomic and phylogenetic studies and was first described as Bacterium vesicatorium as a pathogen of pepper and tomato in 1921.[1] Dowson [2] later reclassified the bacterium as Xanthomonas campestris and proposed the genus Xanthomonas.[3]Xanthomonas was first described as a monotypic genus and further research resulted in the division into two groups, A and B.[4][5] Later work using DNA:DNA hybridization has served as a framework for the general Xanthomonas species classification.[6][7] Other tools, including multilocus sequence analysis and amplified fragment-length polymorphism, have been used for classification within clades.[8][9] While previous research has illustrated the complexity of the Xanthomonas genus, recent research appears to have resulted in a clearer picture. More recently, genome-wide analysis of multiple Xanthomonas strains mostly supports the previous phylogenies.[10]
Morphology and growth
Individual cell characteristics include:
- Cell type – straight rods
- Size – 0.4 – 1.0 µm wide by 1.2 – 3.0 µm long
- Motility – motile by a single polar flagellum
Colony growth characteristics include:
- Mucoid, convex, and yellow colonies on YDC medium [11]
- Yellow pigment from xanthomonadin, which contains bromine
- Most produce large amounts of extracellular polysaccharide
- Temperature range – 4 to 37 °C
Biochemical and physiological test results are:
- Gram stain – negative
- Catalase positive [11]
- Oxidase negative [11]
Xanthomonas plant pathogens
Xanthomonas species can cause bacterial spots and blights of leaves, stems, and fruits on a wide variety of plant species.[12] Pathogenic species show high degrees of specificity and some are split into multiple pathovars, a species designation based on host specificity.
Bacterial blight of cotton, caused by Xanthomonas citri subsp. malvacearum is the most important bacterial disease on cotton which infects all aerial parts of the host. Loss due to this disease was estimated for about 10 to 30% on different cultivars and can be found in Asia, Africa and southern America.[13] Citrus canker, caused by Xanthomonas citri subsp. citri is an economically important disease of many citrus species (lime, orange, lemon, pamelo, etc.)[10]
Bacterial leaf spot has caused significant crop losses over the years. Causes of this disease include Xanthomonas euvesicatoria and Xanthomonas perforans = [Xanthomonas axonopodis (syn. campestris) pv. vesicatoria], Xanthomonas vesicatoria, and Xanthomonas gardneri. In some areas where infection begins soon after transplanting, the total crop can be lost as a result of this disease.[14]
Bacterial blight of rice, caused by Xanthomonas oryzae pv. oryzae, is a disease found worldwide and particularly destructive in the rice-producing regions in Asia.[15]
Plant pathogenesis and disease control
Xanthomonas species can be easily spread in water, movement of infected material such as seed or propagation plants, and by mechanical means such as infected pruning tools. Upon contact with a susceptible host, bacteria enter through wounds or natural plant openings as a means to infect.[14] They inject a number of effector proteins, including TAL effectors, into the plant by their secretion systems (i.e., type III secretion system).
To prevent infections, limiting the introduction of the bacteria is key. Some resistant cultivars of certain plant species are available as this may be the most economical means for controlling this disease. For chemical control, preventative applications are best to reduce the potential for bacterial development. Copper-containing products offer some protection along with field-grade antibiotics such as oxytetracycline, which is labeled for use on some food crops in the United States. Curative applications of chemical pesticides may slow or reduce the spread of the bacterium, but will not cure already diseased plants.[16] It is important to consult chemical pesticide labels when attempting to control bacterial diseases, as different Xanthomonas species can have different responses to these applications. Over-reliance on chemical control methods can also result in the selection of resistant isolates, so these applications should be considered a last resort.
Industrial use
Xanthomonas species produce an extrapolysaccharide called xanthan gum that has a wide range of industrial uses, including foods, petroleum products, and cosmetics.
Xanthomonas resources
Isolates of most species of Xanthomonas are available from the National Collection of Plant Pathogenic Bacteria in the United Kingdom and other international culture collections such as ICMP in New Zealand, CFBP in France, and VKM in Russia. It also can be taken out from MTCC India.
Multiple genomes of Xanthomonas have been sequenced and additional data sets/tools are available at The Xanthomonas Resource.[17]
References
- ↑ Doidge, E.M. (1921). "A tomato canker". Annual Review of Applied Biology. 7: 407–30. doi:10.1111/j.1744-7348.1921.tb05528.x.
- ↑ Dowson, W.J. (1939). "On the systematic position and generic names of the Gram-negative bacterial plant pathogens". Zentralblatt für Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene.: 177–193.
- ↑ Young, J. M.; Dye, D. W.; Bradbury, J. F.; Panagopoulos, C. G.; Robbs, C. F. (1978). "A proposed nomenclature and classification for plant pathogenic bacteria". New Zealand Journal of Agricultural Research. 21 (1): 153–177. doi:10.1080/00288233.1978.10427397. ISSN 0028-8233.
- ↑ Stall, R.E., Beaulieu, C., Egel, D.S. (1994). "Two genetically diverse groups of strains are included in Xanthomonas campestris pv. vesicatoria". Int J Syst Bacteriol. 44: 47–53. doi:10.1099/00207713-44-1-47.
- ↑ Vauterin, L., Swings, J, Kersters, K.; et al. (1990). "Towards an improved taxonomy of Xanthomonas". Int J Syst Bacteriol. 40: 312–316. doi:10.1099/00207713-40-3-312.
- ↑ Rademaker, J.L.W., Louws, F.J., and Schultz, M.H. (2005). "A comprehensive species to strain taxonomic framework for Xanthomonas". Phytopathology. 95: 1098–1111. doi:10.1094/phyto-95-1098.
- ↑ Vauterin, L., Hoste, B., Kersters, K., and Swings, J. (1995). "Reclassification of Xanthomonas". Int J Syst Evol Microbiol. 45: 472–489. doi:10.1099/00207713-45-3-472.
- ↑ Ah-You, N. Gagnevin, L., Grimont, PAD; et al. (2009). "Polyphasic characterization of xanthomonads pathogenic to members of the Anacardiaceae and their relatedness to species of Xanthomonas.". Int J Syst Evol Microbiol. 59: 306–318. doi:10.1099/ijs.0.65453-0.
- ↑ Young, J.M., Wilkie, J.P., Park, D.S., Watson, D.R.W. (2010). "New Zealand strains of plant pathogenic bacteria classified by multi-locus sequence analysis; proposal of Xanthomonas dyei sp. nov.". Plant Pathol. 59: 270–281. doi:10.1111/j.1365-3059.2009.02210.x.
- 1 2 Rodriguez-R LM, Grajales A, Arrieta-Ortiz ML, Salazar C, Restrepo S, Bernal A (2012). "Genomes-based phylogeny of the genus Xanthomonas". BMC Microbiology. 12: 43. doi:10.1186/1471-2180-12-43.
- 1 2 3 Schaad, N.W., Jones, J.B., Chun, W. (2001). "Laboratory Guide for Identification of Plant Pathogenic Bacteria 3rd Ed.": 175–199.
- ↑ Boch J, Bonas U (September 2010). "XanthomonasAvrBs3 Family-Type III Effectors: Discovery and Function". Annual Review of Phytopathology. 48: 419–36. doi:10.1146/annurev-phyto-080508-081936. PMID 19400638.
- ↑ Ali Razaghi, Nader Hasanzadeh, Abolghasem Ghasemi (16 February 2012). "Characterization of Xanthomonas citri subsp. malvacearum strains in Iran" (PDF). African Journal of Microbiology Research. pp. 1165–1170.
- 1 2 Ritchie, D.F (2000). "Bacterial spot of pepper and tomato". The Plant Health Instructor. doi:10.1094/PHI-I-2000-1027-01.
- ↑ Mew, T.W., Alvarez, A.M., Leach, J.E., and Swings, J. (1993). "Focus on bacterial blight of rice". Plant Disease. 77: 5–13. doi:10.1094/pd-77-0005.
- ↑ Ellis, S.D., Boehm, M.J., and Coplin, D. (2008). "Bacterial Diseases of Plants". Ohio State University Fact Sheet.
- ↑ Ralf Koebnik (7 August 2012), The Xanthomonas Resource