Rhizoctonia solani
| Rhizoctonia solani (Anamorph) | |
|---|---|
 | |
| R. solani hyphae showing the distinguishing right angles | |
| Scientific classification | |
| Kingdom: | Fungi |
| Phylum: | Basidiomycota |
| Class: | Agaricomycetes |
| Order: | Cantharellales |
| Family: | Ceratobasidiaceae |
| Genus: | Rhizoctonia |
| Species: | R. solani |
| Binomial name | |
| Rhizoctonia solani J.G. Kühn 1858 | |
| Synonyms | |
|
Moniliopsis aderholdii Ruhland 1908 | |
Rhizoctonia solani (teleomorph: Thanatephorus spp.) is a plant pathogenic fungus with a wide host range and worldwide distribution. This plant pathogen was discovered more than 100 years ago. R. solani frequently exists as thread-like growth on plants or in culture, and is considered a soil-borne pathogen. R. solani is best known to cause various plant diseases such as collar rot, root rot, damping off and wire stem. R. solani attacks its hosts when they are in their juvenile stages of development such as seeds and seedlings, which are typically found in the soil. It makes sense then that this saprophytic pathogen would live and survive in the soil and attack the part of its hosts that reside there. The pathogen is known to cause serious plant losses by attacking primarily the roots and lower stems of plants. Although it has a wide range of hosts, its main targets are herbaceous plants. R. solani would be considered a basidiomycete fungus if the teleomorph stage were more abundant. The pathogen is not currently known to produce any asexual (conidia) spores, though it is considered to have an asexual life cycle. Occasionally, sexual spores (basidiospores) are produced on infected plants. The disease cycle of R. solani is important in regards to management and control of the pathogen.
History
In 1858, Julius Kuhn observed and described a fungus on diseased potato tubers and named it Rhizoctonia solani. Rhizooctonia is from Ancient Greek, ῥίζα (rhiza, "root") + κτόνος (ktonos, "murder"). Solanum, Latin for nightshade, is the genus of the potato. The disease caused was well-known before the discovery and description of the fungus.[1]
Hosts and symptoms
Rhizoctonia solani causes a wide range of commercially significant plant diseases. It is one of the fungi responsible for Brown patch (a turfgrass disease), damping off (e.g. in soybean seedlings),[2] as well as black scurf of potatoes,[3] bare patch of cereals,[4] root rot of sugar beet,[5] belly rot of cucumber,[6] sheath blight of rice,[7] and many other pathogenic conditions. The fungus therefore has a wide host range and strains of R. solani may differ in the hosts they are able to infect, the virulence of infection, selectivity for a given host ranges from non-pathogenic to highly virulent, the temperature at which infection occurs, the ability to develop in lower soil levels, the ability to form sclerotia, the growth rate, the survival in a certain area. These factors may or may not show up given the environment and host that Rhizoctonia attacks.[8]

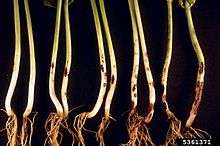
Rhizoctonia solani primarily attacks seeds of plants below the soil surface, but can also infect pods, roots, leaves and stems. The most common symptom of Rhizoctonia is "damping off", or the failure of infected seeds to germinate. R. solani may invade the seed before it has germinated to cause this pre-emergent damping off, or it can kill very young seedlings soon after they emerge from the soil. Seeds that do germinate before being killed by the fungus have reddish-brown lesions and cankers on stems and roots.
There are various environmental conditions that put the plant at higher risk of infection due to Rhizoctonia, the pathogen prefers warmer wet climates for infection and growth. Post-emergent damping off is a further delay in attack of Rhizoctonia solani. The seedling is most susceptible to disease in its juvenile stage.[9]
Cereals in regions of England, South Australia, Canada, and India experience losses caused by Rhizoctonia solani every year. Roots are killed back, causing plants to be stunted and spindly. Other non cereal plants in those regions can experience brown stumps as another symptom of the pathogen. In England, this is called purple patch. R. solani can also cause hypocotyl and stem cankers on mature plants of tomatoes, potatoes and cabbage. Strands of mycelium and sometimes sclerotia appear on their surfaces. Roots will turn brown and die after a period of time. The best known symptom of R. solani is black scurf on potato tubers which are the sclerotia of the fungus.
Disease cycle
Rhizoctonia solani can survive in the soil for many years in the form of sclerotia. Sclerotia of Rhizoctonia have thick outer layers to allow for survival, and they function as the overwintering structure for the pathogen. In some rare cases(such as the teleomorph) the pathogen may also take on the form of mycelium that reside in the soil as well. The fungus is attracted to the plant by chemical stimuli released by a growing plant and/or decomposing plant residue. The process of penetration of a host can be accomplished in a number of ways. Entry can occur through direct penetration of the plant cuticle/epidermis or by means of natural openings in the plant. Hyphae will come in contact with the plant and attach to the plant by which through growth they begin to produce an appressorium which penetrates the plant cell and allows for the pathogen to obtain nutrients from the plant cell. The pathogen can also release enzymes that break down plant cell walls, and continues to colonize and grow inside dead tissue. This breakdown of the cell walls and colonization of the pathogen within the host is what forms the sclerotia. New inoculum is produced on or within the host tissue, and a new cycle is repeated when new plants become available. The disease cycle begins as such:
- the sclerotia/mycelium overwinter in plant debris, soil or host plants.
- The young hyphae and fruiting basidia (rare) emerge and produce mycelia and rarely basidiospores.
- The very rare production of the germinating basidiospores penetrate the stoma whereas the mycelia land on the plant surface and secrete the necessary enzymes onto the plant surface in order to initiate invasion of the host plant.
- After the mycelia successfully invade the host, necrosis and sclerotia form in and around the infected tissue which then leads to the various symptoms associated with the disease such as soil rot, stem rot, damping off etc. and the process begins all over again.[10]
Environment
The pathogen is known to prefer warm wet weather, and outbreaks typically occur in the early summer months. Most symptoms of the pathogen do not occur until late summer and thus most farmers do not become aware of the diseased crop until harvest. A combination of environmental factors have been linked to the prevalence of the pathogen such as: presence of host plant, frequent rainfall/irrigation and increased temperatures in spring and summer. In addition, a reduction of drainage of the soil due to various techniques such as soil compaction are also known to create favorable environments for the pathogen.[11] The pathogen is dispersed as sclerotia, and these sclerotia can travel by means of wind, water or soil movement between host plants.
Identification

| Thanatephorus spp. (Teleomorph) | |
|---|---|
| Scientific classification | |
| Domain: | Eukarya |
| Kingdom: | Fungi |
| Phylum: | Basidiomycota |
| Class: | Agaricomycetes |
| Order: | Cantharellales |
| Family: | Ceratobasidiaceae |
| Genus: | Thanatephorus |
| Species: | Thanatephorus spp. |
| Binomial name | |
| Thanatephorus spp. (A.B. Frank) Donk 1956 | |
| Synonyms | |
|
Corticium sasakii (Shirai) H. Matsumoto 1934 | |
Rhizoctonia solani does not produce spores and is hence identified only from mycelial characteristics or DNA analysis. Its hyphal cells are multinucleate. It produces white to deep brown mycelium when grown on artificial medium. The hyphae are 4–15 μm wide and tend to branch at right angles. A septum near each hyphal branch and a slight constriction at the branch are diagnostic. R. solani is subdivided into anastomosis groups (AG) based on hyphal fusion between compatible strains.[12][13]
The teleomorph of R. solani is Thanatephorus cucumeris. It forms club-shaped basidia with four apical sterigmata on which oval, hyaline basidiospores are borne.
Management
It is not possible to completely control Rhizoctonia, but the severity of the pathogen can be limited. Successful control of Rhizoctonia depends on characteristics of the pathogen, host crops, and environment.[14] Controlling the environment, crop rotation, using resistant varieties, and minimizing soil compaction are effective and noninvasive ways to manage disease. Planting seedlings in warmer soil and getting plants to emerge quickly helps minimize damage. Crop rotation also helps minimize the amount of inoculum that causes Rhizoctonia solani. There are a few resistant varieties with moderate resistance to Rhizoctonia that can be used, but they produce lower yields and quantity than standard varieties. Minimizing soil compaction is also another way to reduce risk of the pathogen because this helps water infiltration, drainage, and aeration for the plants.
One specific chemical option is a chemical spray PCNB which is known to be the best solution to reducing damping off of seeds on host plants. To minimize disease, we can use plant certified seed that is free of sclerotia. Seed growers should look into only purchasing sclerotia free seeds when planting their crops since sclerotia can overwinter in the soil and may not show symptoms right away. Although fungicides are not the most effective way to manage this pathogen, there have been a few that have been approved by the USDA for control of the pathogen. One should consult their chemical representative on which group of fungicides would be most effective with their crops in regard to Rhizoctonia solani.
Avoidance is one of the key ways to ensure that the pathogen will not be among their crops. As long as seed growers stay clear of wet, poorly drained areas while also avoiding susceptible crops, Rhizoctonia solani is not usually a problem. Diseases caused by this pathogen are more severe in soils that are moderately wet and a temperature range of 15-18 degrees C.[15]
Economic Importance
Rhizoctonia can be found across all areas of the United States (environmental conditions permitting) where its host crops are located. The severity of infection can vary and for highly infected patches, severity of the infection can be very devastating to the farmer. Some of these consequences are: major yield losses (ranging from 25%-100%), increased soil tare (because the soil sticks to the fungus' mycelium), and poor industrial quality of the crops based on increased levels of sodium, potassium and nitrogen. Due to the vast number of hosts that the pathogen attacks, these consequences are numerous and detrimental to a variety of crops. Sheath blight caused by this pathogen is the second most devastating disease after rice blast.[16]
Genome
The draft genome of R. solani strain Rhs1AP covers 51.7 Mbp, although the heterokaryotic genome of this strain was estimated at 86 Mb, based on an optical map of the chromosomes. The discrepancy is explained by the aneuploid, highly repetitive genome of this species which prevented sequencing (or assembling) the complete DNA. The genome is predicted to encode 12,726 genes.[17] Another strain, AG1-IB 7/3/14, was recently sequenced too.[18]
References
- ↑ [Parmeter, J. R. Rhizoctonia Solani, Biology and Pathology. London, UK: University of California, 1970. Print.], University of California Biology and Pathology.
- ↑ Shanmugasundaram, S.; Yeh, C.C.; Hartman, G.L.; Talekar, N.S. (1991). Vegetable Soybean Research Needs for Production and Quality Improvement (PDF). Taipei: Asian Vegetable Research and Development Center. pp. 86–87. ISBN 9789290580478. Retrieved 6 February 2016.
- ↑ Rhizoctonia disease of potato http://vegetablemdonline.ppath.cornell.edu/factsheets/Potato_Rhizoctonia.htm
- ↑ Rhizoctonia root rot http://cbarc.aes.oregonstate.edu/rhizoctonia-root-rot-bare-patch
- ↑ Rhizoctonia diseases of sugar beet http://www.uiweb.uidaho.edu/sugarbeet/Disease/rtdspnw.htm
- ↑ Rhizoctonia disease of cucumber http://cuke.hort.ncsu.edu/cucurbit/cuke/dshndbk/br.html
- ↑ Rhizoctonia sheath blight http://www.lsuagcenter.com/NR/rdonlyres/C93A494B-8105-4804-9DFA-81190EC3F68B/58166/pub3123ShealthBlightofRiceHIGHRES.pdf
- ↑ [Ogoshi, Akira. "ECOLOGY AND PATHOGENICITY OF ANASTOMOSIS AND INTRASPECIFIC GROUPS OF .. RHIZOCTONIA SOLANI KUHN." Annual Reviews of Phytopathology 25.125-43 (1987)]
- ↑ [Cubeta, M. A., and R. Vilgalys. "Population Biology of the Rhizoctonia Solani Complex." Population Genetics of Soilborne Fungal Plant Pathogens 87.4 (1997): 480-84. The American Phytopathological Society. Web. 4 Oct. 2011.], The American Phytopathological Society.
- ↑ [Ceresini, Paulo. "Rhizoctonia Solani." Rhizoctonia Solani. NC State University. Web. 04 November 2011 <http://www.cals.ncsu.edu/course/pp728/Rhizoctonia/Rhizoctonia.html>.], NC State University Rhizoctonia Solani.
- ↑ ["Rhizoctonia Diseases." Michigan Potato Diseases. P.S. Wharton, Michigan State University, 2 May 2011. Web. 04 Oct. 2011. <http://www.potatodiseases.org/rhizoctonia.html>.], P.S Wharton Michigan State University.
- ↑ Wiese, M.V. (1987). Compendium of wheat diseases. American Phytopathological Society. pp. 124 pp.
- ↑ APSnet Education Center - Plant Disease Lessons - Rhizoctonia diseases of turfgrass - Pathogen Biology. Plant Pathology / Plant Disease Online - The American Phytopathological Society.
- ↑ [Uchida, Janice Y. "Rhizoctonia Solani." Knowledge Master. Web. 04 Oct. 2011. <http://www.extento.hawaii.edu/kbase/crop/type/r_solani.htm>.], Janice Uchilda Knowledge Master.
- ↑ [Anderson, Neil. "The Genetics and Pathology of Rhizoctonia Solani." Annual Reviews of Phytopathology 20.329-47 (1982): 331-38.]
- ↑ Molecular Plant Pathology (2013) 14(9), 910–922
- ↑ Cubeta MA, Thomas E, Dean RA, Jabaji S, Neate SM, Tavantzis S, Toda T, Vilgalys R, Bharathan N, Fedorova-Abrams N, Pakala SB, Pakala SM, Zafar N, Joardar V, Losada L, Nierman WC (2014). "Draft Genome Sequence of the Plant-Pathogenic Soil Fungus Rhizoctonia solani Anastomosis Group 3 Strain Rhs1AP". Genome Announc. 2 (5): e01072–14. doi:10.1128/genomeA.01072-14. PMC 4214984
 . PMID 25359908.
. PMID 25359908. - ↑ Wibberg D, Rupp O, Jelonek L, Kröber M, Verwaaijen B, Blom J, Winkler A, Goesmann A, Grosch R, Pühler A, Schlüter A (2015). "Improved genome sequence of the phytopathogenic fungus Rhizoctonia solani AG1-IB 7/3/14 as established by deep mate-pair sequencing on the MiSeq (Illumina) system". J. Biotechnol. 203: 19–21. doi:10.1016/j.jbiotec.2015.03.005. PMID 25801332.