Siderophore
Siderophores (Greek: "iron carrier") are small, high-affinity iron chelating compounds secreted by microorganisms such as bacteria, fungi and grasses.[1][2][3][4][5][6] Siderophores are amongst the strongest soluble Fe3+ binding agents known.
The scarcity of soluble iron
Iron is essential for almost all life for processes such as respiration and DNA synthesis. Despite being one of the most abundant elements in the Earth’s crust, the bioavailability of iron in many environments such as the soil or sea is limited by the very low solubility of the Fe3+ ion. This is the predominant state of iron in aqueous, non-acidic, oxygenated environments. It accumulates in common mineral phases such as iron oxides and hydroxides (the minerals that are responsible for red and yellow soil colours) hence cannot be readily used by organisms.[7] Microbes release siderophores to scavenge iron from these mineral phases by formation of soluble Fe3+ complexes that can be taken up by active transport mechanisms. Many siderophores are nonribosomal peptides,[3][8] although several are biosynthesised independently.[9]
Siderophores are also important for some pathogenic bacteria for their acquisition of iron.[3][4][6] In mammalian hosts, iron is tightly bound to proteins such as hemoglobin, transferrin, lactoferrin and ferritin. The strict homeostasis of iron leads to a free concentration of about 10−24 mol L−1,[10] hence there are great evolutionary pressures put on pathogenic bacteria to obtain this metal. For example, the anthrax pathogen Bacillus anthracis releases two siderophores, bacillibactin and petrobactin, to scavenge ferric iron from iron proteins. While bacillibactin has been shown to bind to the immune system protein siderocalin,[11] petrobactin is assumed to evade the immune system and has been shown to be important for virulence in mice.[12]
Siderophores are amongst the strongest binders to Fe3+ known, with enterobactin being one of the strongest of these.[10] Because of this property, they have attracted interest from medical science in metal chelation therapy, with the siderophore desferrioxamine B gaining widespread use in treatments for iron poisoning and thalassemia.[13]
Besides siderophores, some pathogenic bacteria produce hemophores (heme binding scavenging proteins) or have receptors that bind directly to iron/heme proteins.[14] In eukaryotes, other strategies to enhance iron solubility and uptake are the acidification of the surroundings (e.g. used by plant roots) or the extracellular reduction of Fe3+ into the more soluble Fe2+ ions.
Structure

Siderophores usually form a stable, hexadentate, octahedral complex preferentially with Fe3+ compared to other naturally occurring abundant metal ions, although if there are fewer than six donor atoms water can also coordinate. The most effective siderophores are those that have three bidentate ligands per molecule, forming a hexadentate complex and causing a smaller entropic change than that caused by chelating a single ferric ion with separate ligands.[15] A comprehensive list of siderophores is presented in.[16] Fe3+ is a hard Lewis acid, preferring hard Lewis bases such as anionic or neutral oxygen atoms to coordinate with. Microbes usually release the iron from the siderophore by reduction to Fe2+ which has little affinity to these ligands.[8][17]
Siderophores are usually classified by the ligands used to chelate the ferric iron. The major groups of siderophores include the catecholates (phenolates), hydroxamates and carboxylates (e.g. derivatives of citric acid).[3] Citric acid can also act as a siderophore.[18] The wide variety of siderophores may be due to evolutionary pressures placed on microbes to produce structurally different siderophores which cannot be transported by other microbes' specific active transport systems, or in the case of pathogens deactivated by the host organism.[3][6]
Diversity
Examples of siderophores produced by various bacteria and fungi:
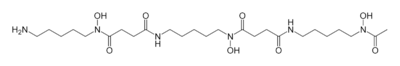

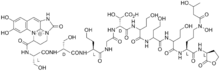


Hydroxamate siderophores
| Siderophore | Organism |
|---|---|
| ferrichrome | Ustilago sphaerogena |
| Desferrioxamine B | Streptomyces pilosus |
| Desferrioxamine E | Streptomyces coelicolor |
| fusarinine C | Fusarium roseum |
| ornibactin | Burkholderia cepacia |
| rhodotorulic acid | Rhodotorula pilimanae |
Catecholate siderophores
| Siderophore | Organism |
|---|---|
| enterobactin | Escherichia coli
enteric bacteria |
| bacillibactin | Bacillus subtilis |
| vibriobactin | Vibrio cholerae |
Mixed ligands
| Siderophore | Organism |
|---|---|
| azotobactin | Azotobacter vinelandii |
| pyoverdine | Pseudomonas aeruginosa |
| yersiniabactin | Yersinia pestis |
A comprehensive list of siderophore structures (over 250) is presented in Appendix 1 in reference.[3]
Biological function
Bacteria and fungi
In response to iron limitation in their environment, genes involved in microbe siderophore production and uptake are derepressed, leading to manufacture of siderophores and the appropriate uptake proteins. In bacteria, Fe2+-dependent repressors bind to DNA upstream to genes involved in siderophore production at high intracellular iron concentrations. At low concentrations, Fe2+ dissociates from the repressor, which in turn dissociates from the DNA, leading to transcription of the genes. In gram-negative and AT-rich gram-positive bacteria, this is usually regulated by the Fur (ferric uptake regulator) repressor, whilst in GC-rich gram-positive bacteria (e.g. Actinobacteria) it is DtxR (diphtheria toxin repressor), so-called as the production of the dangerous diphtheria toxin by Corynebacterium diphtheriae is also regulated by this system.[8]
This is followed by excretion of the siderophore into the extracellular environment, where the siderophore acts to sequester and solubilize the iron.[3][19][20][21] Siderophores are then recognized by cell specific receptors on the outer membrane of the cell.[2][3][22] In fungi and other eukaryotes, the Fe-siderophore complex may be extracellularly reduced to Fe2+, while in many cases the whole Fe-siderophore complex is actively transported across the cell membrane. In gram-negative bacteria, these are transported into the periplasm via TonB-dependent receptors, and are transferred into the cytoplasm by ABC transporters.[3][8][15][23]
Once in the cytoplasm of the cell, the Fe3+-siderophore complex is usually reduced to Fe2+ to release the iron, especially in the case of “weaker” siderophore ligands such as hydroxamates and carboxylates. Siderophore decomposition or other biological mechanisms can also release iron.,[15] especially in the case of catecholates such as ferric-enterobactin, whose reduction potential is too low for reducing agents such as flavin adenine dinucleotide, hence enzymatic degradation is needed to release the iron.[10]
Plants
Although there is sufficient iron in most soils for plant growth, plant iron deficiency is a problem in calcareous soil, due to the low solubility of iron(III) hydroxide. Calcareous soil accounts for 30% of the world's farmland. Under such conditions graminaceous plants (grasses, cereals and rice) secrete phytosiderophores into the soil,[24] a typical example being deoxymugineic acid. Phytosiderophores have a different structure to those of fungal and bacterial siderophores having two α-aminocarboxylate binding centres, together with a single α-hydroxycarboxylate unit. This latter bidentate function provides phytosiderophores with a high selectivity for iron(III). When grown in an iron -deficient soil, roots of graminaceous plants secrete siderophores into the rhizosphere. On scavenging iron(III) the iron –phytosiderophore complex is transported across the cytoplasmic membrane using a proton symport mechanism.[25] The iron(III) complex is then reduced to iron(II) and the iron is transferred to nicotianamine, which although very similar to the phytosiderophores is selective for iron(II) and is not secreted by the roots.[26] Nicotianamine translocates iron in phloem to all plant parts.
Siderophore ecology
Siderophores become important in the ecological niche defined by low iron availability, iron being one of the critical growth limiting factors for virtually all aerobic microorganisms. There are four major ecological habitats: soil and surface water, marine water, plant tissue (pathogens) and animal tissue (pathogens).
Soil and surface water
The soil is a rich source of bacterial and fungal genera. Common Gram-positive species are those belonging to the Actinomycetales and species of the genera Bacillus, Arthrobacter and Nocardia. Many of these organisms produce and secrete ferrioxamines which lead to growth promotion of not only the producing organisms, but also other microbial populations that are able to utilize exogenous siderophores . Soil fungi include Aspergillus and Penicillium which predominately produce ferrichromes. This group of siderophores consist of cyclic hexapeptides and consequently are highly resistant to environmental degradation associated with the wide range of hydrolytic enzymes that are present in humic soil.[27] Soils containing decaying plant material possess pH values as low as 3–4. Under such conditions organisms that produce hydroxamate siderophores have an advantage due to the extreme acid stability of these molecules. The microbial population of fresh water is similar to that of soil, indeed many bacteria are washed out from the soil. In addition, fresh-water lakes contain large populations of Pseudomonas, Azomonas, Aeromonos and Alcaligenes species.[28]
Marine water
In contrast to most fresh-water sources, iron levels in surface sea-water are extremely low (1 nM to 1 μM in the upper 200 m) and much lower than those of V, Cr, Co, Ni, Cu and Zn. Virtually all this iron is in the iron(III) state and complexed to organic ligands.[29] These low levels of iron limit the primary production of phytoplankton and have led to the Iron Hypothesis[30] where it was proposed that an influx of iron would promote phytoplankton growth and thereby reduce atmospheric CO2. This hypothesis has been tested on more than 10 different occasions and in all cases, massive blooms resulted. However, the blooms persisted for variable periods of time. An interesting observation made in some of these studies was that the concentration of the organic ligands increased over a short time span in order to match the concentration of added iron, thus implying biological origin and in view of their affinity for iron possibly being of a siderophore or siderophore-like nature.[31] Significantly, heterotrophic bacteria were also found to markedly increase in number in the iron-induced blooms. Thus there is the element of synergism between phytoplankton and heterotrophic bacteria. Phytoplankton require iron (provided by bacterial siderophores), and heterotrophic bacteria require non-CO2 carbon sources (provided by phytoplankton).
The dilute nature of the pelagic marine environment promotes large diffusive losses and renders the efficiency of the normal siderophore-based iron uptake strategies problematic. However, many heterotrophic marine bacteria do produce siderophores, albeit with properties different from those produced by terrestrial organisms. Many marine siderophores are surface-active and tend to form molecular aggregates, for example aquachelins. The presence of the fatty acyl chain renders the molecules with a high surface activity and an ability to form micelles.[32] Thus, when secreted, these molecules bind to surfaces and to each other, thereby slowing the rate of diffusion away from the secreting organism and maintaining a relatively high local siderophore concentration. Phytoplankton have high iron requirements and yet the majority (and possibly all) do not produce siderophores . Phytoplankton can, however, obtain iron from siderophore complexes by the aid of membrane-bound reductases [33] and certainly from iron(II) generated via photochemical decomposition of iron(III) siderophores. Thus a large proportion of iron (possibly all iron) absorbed by phytoplankton is dependent on bacterial siderophore production.
[34] Plant pathogens
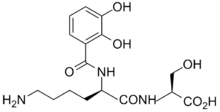
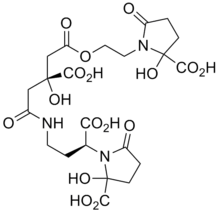
Most plant pathogens invade the apoplasm by releasing pectolytic enzymes which facilitate the spread of the invading organism. Bacteria frequently infect plants by gaining entry to the tissue via the stomata. Having entered the plant they spread and multiply in the intercellular spaces. With bacterial vascular diseases, the infection is spread within the plants through the xylem.
Once within the plant, the bacteria need to be able to scavenge iron from the two main iron-transporting ligands, nicotianamine and citrate.[35] To do this they produce siderophores, thus the enterobacterial Erwinia chrysanthemi produces two siderophores, chrysobactin and achromobactin.[36]
Similar like in humans also plants possess siderophore binding proteins involve in host defense like the major birch pollen allergen, Bet v 1, which are usually secreted and possess a lipocalin-like structure.[34]
Animal pathogens
Pathogenic bacteria and fungi have developed the means of survival in animal tissue. They may invade the gastro-intestinal tract (Escherichia, Shigella and Salmonella), the lung (Pseudomonas, Bordatella, Streptococcus and Corynebacterium), skin (Staphylococcus) or the urinary tract (Escherichia and Pseudomonas). Such bacteria may colonise wounds (Vibrio and Staphylococcus) and be responsible for septicaemia (Yersinia and Bacillus). Some bacteria survive for long periods of time in intracellular organelles, for instance Mycobacterium. (see table). Because of this continual risk of bacterial and fungal invasion, animals have developed a number of lines of defence based on immunological strategies, the complement system, the production of iron–siderophore binding proteins and the general “withdrawal”of iron.[37]
| Infection type | Organism | Siderophore |
|---|---|---|
| Dysentery | Shigella sp. | Aerobactin |
| Intestinal infections | Escherichia coli | Enterobactin |
| Typhoid | Salmonella sp. | Salmochelin |
| Plague | Yersinia sp. | Yersiniabactin |
| Cholera | Vibrio sp. | Vibriobactin |
| Pulmonary infections | Pseudomonas sp. | Pyoverdins |
| Whooping cough | Bordetella sp. | Alcaligin |
| Tuberculosis | Mycobacterium tuberculosis | Mycobactins |
| Skin and mucous membrane infections | Staphylococcus sp. | Staphyloferrin A |
| Anthrax | Bacillus anthracis | Petrobactin |
There are two major types of iron-binding proteins present in most animals that provide protection against microbial invasion – extracellular protection is achieved by the transferrin family of proteins and intracellular protection is achieved by ferritin. Transferrin is present in the serum at approximately 30 μM, and contains two iron-binding sites, each with an extremely high affinity for iron. Under normal conditions it is about 25–40% saturated, which means that any freely available iron in the serum will be immediately scavenged – thus preventing microbial growth . Most siderophores are unable to remove iron from transferrin. Mammals also produce lactoferrin, which is similar to serum transferrin but possesses an even higher affinity for iron.[38] Lactoferrin is present in secretory fluids, such as sweat, tears and milk, thereby minimising bacterial infection.
Ferritin is present in the cytoplasm of cells and limits the intracellular iron level to approximately 1 μM. Ferritin is a much larger protein than transferrin and is capable of binding several thousand iron atoms in a nontoxic form. Siderophores are unable to directly mobilise iron from ferritin.
In addition to these two classes of iron-binding proteins, a hormone, hepcidin, is involved in controlling the release of iron from absorptive enterocytes, iron-storing hepatocytes and macrophages.[39] Infection leads to inflammation and the release of interleukin-6 (IL-6 ) which stimulates hepcidin expression. In humans, IL-6 production results in low serum iron, making it difficult for invading pathogens to infect. Such iron depletion has been demonstrated to limit bacterial growth in both extracellular and intracellular locations.[37]
In addition to “iron withdrawal” tactics, mammals produce an iron –siderophore binding protein, siderochelin. Siderochelin is a member of the lipocalin family of proteins, which while diverse in sequence, displays a highly conserved structural fold, an 8-stranded antiparallel β-barrel that forms a binding site with several adjacent β-strands. Siderocalin (lipocalin 2) has 3 positively charged residues also located in the hydrophobic pocket, and these create a high affinity binding site for iron(III)–enterobactin.[40] Siderocalin is a potent bacteriostatic agent against E. coli. As a result of infection it is secreted by both macrophages and hepatocytes, enterobactin being scavenged from the extracellular space.
Medical applications
Siderophores have applications in medicine for iron and aluminum overload therapy and antibiotics for improved targeting.[3] Understanding the mechanistic pathways of siderophores has led to opportunities for designing small-molecule inhibitors that block siderophore biosynthesis and therefore bacterial growth and virulence in iron-limiting environments.[41]
Siderophores are useful as drugs in facilitating iron mobilization in humans, especially in the treatment of iron diseases, due to their high affinity for iron. One potentially powerful application is to use the iron transport abilities of siderophores to carry drugs into cells by preparation of conjugates between siderophores and antimicrobial agents. Because microbes recognize and utilize only certain siderophores, such conjugates are anticipated to have selective antimicrobial activity.[6][15]
Microbial iron transport (siderophore)-mediated drug delivery makes use of the recognition of siderophores as iron delivery agents in order to have the microbe assimilate siderophore conjugates with attached drugs. These drugs are lethal to the microbe and cause the microbe to apoptosise when it assimilates the siderophore conjugate.[6] Through the addition of the iron-binding functional groups of siderophores into antibiotics, their potency has been greatly increased. This is due to the siderophore-mediated iron uptake system of the bacteria.
Agricultural applications
Poaceae (grasses) including agriculturally important species such as barley and wheat are able to efficiently sequester iron by releasing phytosiderophores via their root into the surrounding soil rhizosphere.[19] Chemical compounds produced by microorganisms in the rhizosphere can also increase the availability and uptake of iron. Plants such as oats are able to assimilate iron via these microbial siderophores. It has been demonstrated that plants are able to use the hydroxamate-type siderophores ferrichrome, rodotorulic acid and ferrioxamine B; the catechol-type siderophores, agrobactin; and the mixed ligand catechol-hydroxamate-hydroxy acid siderophores biosynthesized by saprophytic root-colonizing bacteria. All of these compounds are produced by rhizospheric bacterial strains, which have simple nutritional requirements, and are found in nature in soils, foliage, fresh water, sediments, and seawater.[42]
Fluorescent pseudomonads have been recognized as biocontrol agents against certain soil-borne plant pathogens. They produce yellow-green pigments (pyoverdines) which fluoresce under UV light and function as siderophores. They deprive pathogens of the iron required for their growth and pathogenesis.[43]
Other metals chelated
Siderophores can chelate metals other than iron. Examples include aluminium,[2][22][42][44] gallium,[2][22][42][44] chromium,[22][42] copper,[22][42][44] zinc,[22][44] lead,[22] manganese,[22] cadmium,[22] vanadium,[22] indium,[22][44] plutonium,[45] and uranium.[45]
Related processes
Alternative means of assimilating iron are surface reduction, lowering of pH, utilization of heme, or extraction of protein-complexed metal.[2]
See also
References
- ↑ J. B. Neilands (1952). "A Crystalline Organo-iron Pigment from a Rust Fungus (Ustilago sphaerogena)". J. Am. Chem. Soc. 74 (19): 4846–4847. doi:10.1021/ja01139a033.
- 1 2 3 4 5 J. B. Neilands (1995). "Siderophores: Structure and Function of Microbial Iron Transport Compounds". J. Biol. Chem. 270 (45): 26723–26726. doi:10.1074/jbc.270.45.26723. PMID 7592901.
- 1 2 3 4 5 6 7 8 9 10 R. C. Hider & X. Kong (2010). "Chemistry and biology of siderophores". Nat. Prod. Rep. 27: 637–657. doi:10.1039/b906679a.
- 1 2 J. H. Crosa, A. R. Mey, S. M. Payne (editor) (2004). Iron Transport in Bacteria. ASM Press. ISBN 1-55581-292-9.
- ↑ Cornelis, P; Andrews, SC (editor) (2010). Iron Uptake and Homeostasis in Microorganisms. Caister Academic Press. ISBN 978-1-904455-65-3.
- 1 2 3 4 5 Miller, Marvin J. Siderophores (microbial iron chelators) and siderophore-drug conjugates (new methods for microbially selective drug delivery). University of Notre Dame, 4/21/2008 http://www.nd.edu/~mmiller1/page2.html
- ↑ Kraemer, Stephan M. (2005). "Iron oxide dissolution and solubility in the presence of siderophores". Aquatic Sciences. 66: 3–18. doi:10.1007/s00027-003-0690-5.
- 1 2 3 4 Miethke, M.; Marahiel, M. (2007). "Siderophore-Based Iron Acquisition and Pathogen Control". Microbiology and Molecular Biology Reviews. 71 (3): 413–451. doi:10.1128/MMBR.00012-07. PMC 2168645
 . PMID 17804665.
. PMID 17804665. - ↑ Challis, G. L. (2005). "A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases". ChemBioChem. 6 (4): 601–611. doi:10.1002/cbic.200400283. PMID 15719346.
- 1 2 3 Raymond, K. N.; Dertz, E. A.; Kim, S. S. (2003). "Enterobactin: An archetype for microbial iron transport". PNAS. 100 (7): 3584–3588. doi:10.1073/pnas.0630018100. PMC 152965
 . PMID 12655062.
. PMID 12655062. - ↑ Rebecca J. Abergel; Melissa K. Wilson; Jean E. L. Arceneaux; Trisha M. Hoette; Roland K. Strong; B. Rowe Byers & Kenneth N. Raymond (2006). "Anthrax pathogen evades the mammalian immune system through stealth siderophore production". PNAS. 103 (49): 18499–18503. doi:10.1073/pnas.0607055103. PMC 1693691
 . PMID 17132740.
. PMID 17132740. - ↑ Cendrowski, S., W. MacArthur, and P. Hanna. (2004). "Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence". Molecular Microbiology. 51 (2): 407–417. doi:10.1046/j.1365-2958.2003.03861.x. PMID 14756782.
- ↑ T. Zhou, Y. Ma, X. Kong and R. C. Hider (2012). "Design of iron chelators with therapeutic application.". Dalton Trans. 41 (21): 6371–6389. doi:10.1039/c2dt12159j. PMID 22391807.
- ↑ Krewulak, K. D.; Vogel, H. J. (2008). "Structural biology of bacterial iron uptake". Biochim. Biophys. Acta. 1778 (9): 1781–1804. doi:10.1016/j.bbamem.2007.07.026. PMID 17916327.
- 1 2 3 4 John M. Roosenberg II, Yun-Ming Lin, Yong Lu and Marvin J. Miller (2000). "Studies and Syntheses of Siderophores, Microbial Iron Chelators, and Analogs as Potential Drug Delivery Agents". Current Medicinal Chemistry. 7 (2): 159–197. doi:10.2174/0929867003375353. PMID 10637361.
- ↑ R. C. Hider & X. Kong (2010). "Chemistry and biology of siderophores (Appendix(i) List of siderophorestructures)". Nat. Prod. Rep. 27: 637–657. doi:10.1039/b906679a.
- ↑ Neilands, J. B. Siderophores: Structure and Function of Microbial Iron Transport Compounds. J. Biol. Chem. 1995, 270 (45), 26723–26726.
- ↑ Winkelmann, G.; Drechsel, H., Biotechnology (2nd edition), Chapter 5: Microbial Siderophores. 1999.
- 1 2 Kraemer, Stephan M., Crowley, David, and Kretzschmar, Ruben (2006). "Siderophores in Plant Iron Acquisition: Geochemical Aspects". Advances in Agronomy. Advances in Agronomy. 91: 1–46. doi:10.1016/S0065-2113(06)91001-3. ISBN 978-0-12-000809-4.
- ↑ Kraemer, Stephan M., Butler, Allison, Borer, Paul, and Cervini-Silva, Javiera (2005). "Siderophores and the dissolution of iron bearing minerals in marine systems". Reviews in Mineralogy and Geochemistry. 59: 53–76. doi:10.2138/rmg.2005.59.4.
- ↑ Huyer, Marianne & Page, William J. (1988). "Zn2+ Increases Siderophore Production in Azotobacter vinelandii". Applied and Environmental Microbiology. 54: 2625–2631.
- 1 2 3 4 5 6 7 8 9 10 11 A. del Olmo; C. Caramelo & C. SanJose (2003). "Fluorescent complex of pyoverdin with aluminum". J. Inorg. Biochem. 97 (4): 384–387. doi:10.1016/S0162-0134(03)00316-7. PMID 14568244.
- ↑ David Cobessi; Ahmed Meksem & Karl Brillet (2010). "Structure of the heme/hemoglobin outer membrane receptor ShuA from Shigella dysenteriae: Heme binding by an induced fit mechanism". Proteins: Structure, Function, and Bioinformatics. 78 (2): 286–294. doi:10.1002/prot.22539. PMID 19731368.
- ↑ Y. Sugiura & K. Nomoto (1984). "Phytosiderophores structures and properties of mugineic acids and their metal complexes". Structure and Bonding. 58: 107–135. doi:10.1007/BFb0111313.
- ↑ S. Mori; Sigel, A. and Sigel, H. (editor) (1998). Iron transport in graminaceous plants. Metal Ions in Biological Systems. pp. 216–238.
- ↑ E. L. Walker & E. L. Connolly (2008). "Time to pump iron: iron-deficiency-signaling mechanisms of higher plants". Current Opinion in Plant Biology. 11: 530–535. doi:10.1016/j.pbi.2008.06.013. PMID 18722804.
- ↑ G. Winkelmann (2007). "Ecology of siderophores with special reference to the fungi". BioMetals. 20 (3-4): 379–392. doi:10.1007/s10534-006-9076-1. PMID 17235665.
- ↑ G. Winkelmann; Crosa, J. H. Mey, A. R. and Payne, S. M. (editor) (2004). "28". Iron transport in Bacteria. ASM press. pp. 437–450. ISBN 1-55581-292-9.
- ↑ E. L. Rue & K. W. Bruland (1995). "Complexation of iron(III) by natural organic ligands in the Central North Pacific as determined by a new competitive ligand equilibration/adsorptive cathodic stripping voltammetric method". Mar. Chem. 50: 117–138. doi:10.1016/0304-4203(95)00031-L.
- ↑ J. H. Martin (1990). "Glacial-interglacial CO2 change: The Iron Hypothesis". Paleoceanography. 5: 1–13. Bibcode:1990PalOc...5....1M. doi:10.1029/PA005i001p00001.
- ↑ A. Butler (2005). "Marine siderophores and microbial iron mobilization.". BioMetals. 18 (4): 369–374. doi:10.1007/s10534-005-3711-0. PMID 16158229.
- ↑ G. Xu, J. S. Martinez, J. T. Groves and A. Butler (2002). "Membrane affinity of the amphiphilic marinobactin siderophores.". J. Am. Chem. Soc. 124 (45): 13408–13415. doi:10.1021/ja026768w. PMID 12418892.
- ↑ B. M. Hopkinson & F. M. M. Morel (2009). "The role of siderophores in iron acquisition by photosynthetic marine microorganisms.". BioMetals. 22: 659–669. doi:10.1007/s10534-009-9235-2. PMID 19343508.
- 1 2 Roth-Walter, Franziska; Gomez-Casado, Cristina; Pacios, Luis F.; Mothes-Luksch, Nadine; Roth, Georg A.; Singer, Josef; Diaz-Perales, Araceli; Jensen-Jarolim, Erika (2014-06-20). "Bet v 1 from birch pollen is a lipocalin-like protein acting as allergen only when devoid of iron by promoting Th2 lymphocytes". The Journal of Biological Chemistry. 289 (25): 17416–17421. doi:10.1074/jbc.M114.567875. ISSN 1083-351X. PMC 4067174
 . PMID 24798325.
. PMID 24798325. - ↑ N. von Wiren, S. Klair, S. Bansal, J. -F. Briat, H. Khodr, T. Shioiri, R. A. Leigh and R. C. Hider (1999). "Nicotianamine Chelates Both FeIII and FeII. Implications for Metal Transport in Plants.". Plant Physiol. 119 (3): 1107–1114. doi:10.1104/pp.119.3.1107. PMID 10069850.
- ↑ D. Expert, L. Rauscher and T. Franza; Crosa, J. H. Mey, A. R. and Payne, S. M. (editor) (2004). "26". Iron transport in Bacteria. ASM press. pp. 402–412. ISBN 1-55581-292-9.
- 1 2 E. D. Weinberg (2009). "Iron availability and infection.". Biochim. Biophys. Acta. 1790: 600–605. doi:10.1016/j.bbagen.2008.07.002. PMID 18675317.
- ↑ R. Crichton; Crichton, R. (editor) (2001). Inorganic Biochemistry of Iron Metabolism. Wiley. ISBN 0-471-49223-X.
- ↑ S. Rivera, L. Liu, E. Nemeth, V. Gabayan, O. E. Sorensen and T. Ganz (2005). "Hepcidin excess induces the sequestration of iron and exacerbates tumor-associated anemia.". Blood. 105: 1797–1802. doi:10.1182/blood-2004-08-3375. PMID 15479721.
- ↑ K. N. Raymond, E. A. Dertz and S. S. Kim (2003). "Enterobactin: an archetype for microbial iron transport.". Proc. Natl. Acad. Sci. U.S.A. 100: 3584–3588. doi:10.1073/pnas.0630018100. PMC 152965
 . PMID 12655062.
. PMID 12655062. - ↑ Julian A Ferreras, Jae-Sang Ryu, Federico Di Lello, Derek S Tanand Luis E N Quadri (2005). "Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis". Nature Chemical Biology. 1 (1): 29–32. doi:10.1038/nchembio706. PMID 16407990.
- 1 2 3 4 5 G. Carrillo-Castañeda, J. Juárez Muños, J. R. Peralta-Videa, E. Gomez, K. J. Tiemannb, M. Duarte-Gardea and J. L. Gardea-Torresdey (2002). "Alfalfa growth promotion by bacteria grown under iron limiting conditions". Advances in Environmental Research. 6 (3): 391–399. doi:10.1016/S1093-0191(02)00054-0.
- ↑ K. S. Jagadeesh, J. H. Kulkarni and P. U. Krishnaraj (2001). "Evaluation of the role of fluorescent siderophore in the biological control of bacterial wilt in tomato using Tn5 mutants of fluorescent Pseudomonas sp". Current Science. 81: 882.
- 1 2 3 4 5 R. C. Hider & A. D. Hall (1991). "Clinically useful chelators of tripositive elements.". Prog. Med. Chem. 28: 41–137. doi:10.1016/s0079-6468(08)70363-1. PMID 1843549.
- 1 2 John, Seth G., Ruggiero, Christy E., Hersman, Larry E., Tung, Chang-Shung., and Neu, Mary P. (2001). "Siderophore Mediated Plutonium Accumulation by Microbacterium flavescens (JG-9)". Environ. Sci. Technol. 35 (14): 2942–2948. doi:10.1021/es010590g. PMID 11478246.