Whole genome sequencing

Whole genome sequencing (also known as WGS, full genome sequencing, complete genome sequencing, or entire genome sequencing) is a laboratory process that determines the complete DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondria and, for plants, in the chloroplast.
Whole genome sequencing should not be confused with DNA profiling, which only determines the likelihood that genetic material came from a particular individual or group, and does not contain additional information on genetic relationships, origin or susceptibility to specific diseases.[2] Also unlike full genome sequencing, SNP genotyping covers less than 0.1% of the genome. Almost all truly complete genomes are of microbes; the term "full genome" is thus sometimes used loosely to mean "greater than 95%". The remainder of this article focuses on nearly complete human genomes.
High-throughput genome sequencing technologies have largely been used as a research tool and are currently being introduced in the clinics.[3][4][5] In the future of personalized medicine, whole genome sequence data will be an important tool to guide therapeutic intervention.[6] The tool of gene sequencing at SNP level is also used to pinpoint functional variants from association studies and improve the knowledge available to researchers interested in evolutionary biology, and hence may lay the foundation for predicting disease susceptibility and drug response.
History
.jpg)



The shift from manual DNA sequencing methods such as Maxam-Gilbert sequencing and Sanger sequencing in the 1970s and 1980s to more rapid, automated sequencing methods in the 1990s played a crucial role in giving scientists the ability to sequence whole genomes.[8] Haemophilus influenzae, a commensal bacterium which resides in the human respiratory tract was the first organism to have its entire genome sequenced (Figure 2.1). The entire genome of this bacterium was published in 1995.[9] The genomes of H. influenzae, other Bacteria, and some Archaea were the first to be sequenced - largely due to their small genome size. H. influenzae has a genome of 1,830,140 base pairs of DNA.[9] In contrast, eukaryotes, both unicellular and multicellular such as Amoeba dubia and humans (Homo sapiens) respectively, have much larger genomes (see C-value paradox).[10] Amoeba dubia has a genome of 700 billion nucleotide pairs spread across thousands of chromosomes.[11] Humans contain fewer nucleotide pairs (about 3.2 billion in each germ cell - note the exact size of the human genome is still being revised) than A. dubia however their genome size far outweighs the genome size of individual bacteria.[12]
The first bacterial and archaeal genomes, including that of H. influenzae, were sequenced by Shotgun sequencing.[9] In 1996, the first eukaryotic genome ( the yeast Saccharomyces cerevisiae) was sequenced. S. cerevisiae, a model organism in biology has a genome of only around 12 million nucleotide pairs,[13] and was the first unicellular eukaryote to have its whole genome sequenced. The first multicellular eukaryote, and animal, to have its whole genome sequenced was the nematode worm: Caenorhabditis elegans in 1998.[14] Eukaryotic genomes are sequenced by several methods including Shotgun sequencing of short DNA fragments and sequencing of larger DNA clones from DNA libraries (see library (biology)) such as Bacterial artificial chromosomes (BACs) and Yeast artificial chromosomes (YACs).[15]
In 1999, the entire DNA sequence of human chromosome 22, the shortest human autosome, was published.[16] By the year 2000, the second animal and second invertebrate (yet first insect) genome was sequenced - that of the fruit fly Drosophila melanogaster - a popular choice of model organism in experimental research.[17] The first plant genome - that of the model organism Arabidopsis thaliana - was also fully sequenced by 2000.[18] By 2001, a draft of the entire human genome sequence was published.[19] The genome of the laboratory mouse Mus musculus was completed in 2002.[20]
In 2004, the Human Genome Project published the human genome.[21]
Currently, thousands of genomes have been sequenced.
Experimental details
Cells used for sequencing
Almost any biological sample containing a full copy of the DNA—even a very small amount of DNA or ancient DNA—can provide the genetic material necessary for full genome sequencing. Such samples may include saliva, epithelial cells, bone marrow, hair (as long as the hair contains a hair follicle), seeds, plant leaves, or anything else that has DNA-containing cells.
The genome sequence of a single cell selected from a mixed population of cells can be determined using techniques of single cell genome sequencing. This has important advantages in environmental microbiology in cases where a single cell of a particular microorganism species can be isolated from a mixed population by microscopy on the basis of its morphological or other distinguishing characteristics. In such cases the normally necessary steps of isolation and growth of the organism in culture may be omitted, thus allowing the sequencing of a much greater spectrum of organism genomes.[22]
Single cell genome sequencing is being tested as a method of preimplantation genetic diagnosis, wherein a cell from the embryo created by in vitro fertilization is taken and analyzed before embryo transfer into the uterus.[23] After implantation, cell-free fetal DNA can be taken by simple venipuncture from the mother and used for whole genome sequencing of the fetus.[24]
Early techniques

Sequencing of nearly an entire human genome was first accomplished in 2000 partly through the use of shotgun sequencing technology. While full genome shotgun sequencing for small (4000–7000 base pair) genomes was already in use in 1979,[25] broader application benefited from pairwise end sequencing, known colloquially as double-barrel shotgun sequencing. As sequencing projects began to take on longer and more complicated genomes, multiple groups began to realize that useful information could be obtained by sequencing both ends of a fragment of DNA. Although sequencing both ends of the same fragment and keeping track of the paired data was more cumbersome than sequencing a single end of two distinct fragments, the knowledge that the two sequences were oriented in opposite directions and were about the length of a fragment apart from each other was valuable in reconstructing the sequence of the original target fragment.
The first published description of the use of paired ends was in 1990 as part of the sequencing of the human HPRT locus,[26] although the use of paired ends was limited to closing gaps after the application of a traditional shotgun sequencing approach. The first theoretical description of a pure pairwise end sequencing strategy, assuming fragments of constant length, was in 1991.[27] In 1995 the innovation of using fragments of varying sizes was introduced,[28] and demonstrated that a pure pairwise end-sequencing strategy would be possible on large targets. The strategy was subsequently adopted by The Institute for Genomic Research (TIGR) to sequence the entire genome of the bacterium Haemophilus influenzae in 1995,[29] and then by Celera Genomics to sequence the entire fruit fly genome in 2000,[30] and subsequently the entire human genome. Applied Biosystems, now called Life Technologies, manufactured the automated capillary sequencers utilized by both Celera Genomics and The Human Genome Project.
Current techniques
While capillary sequencing was the first approach to successfully sequence a nearly full human genome, it is still too expensive and takes too long for commercial purposes. Since 2005 capillary sequencing has been progressively displaced by next-generation sequencing technologies such as Illumina dye sequencing, pyrosequencing, and SMRT sequencing.[31] All of these technologies continue to employ the basic shotgun strategy, namely, parallelization and template generation via genome fragmentation.
Other technologies are emerging, including nanopore technology. Though nanopore sequencing technology is still being refined, its portability and potential capability of generating long reads are of relevance to whole-genome sequencing applications.[32]
Analysis
In principle, full genome sequencing can provide raw data on all six billion nucleotides in an individual's DNA. However, it does not provide an analysis of what that information means or how it might be utilized in various clinical applications, such as in medicine to help prevent disease. Work toward that goal is continuously moving forward.
Because sequencing generates a lot of data (for example, there are approximately six billion base pairs in each human diploid genome), its output is stored electronically and requires a large amount of computing power and storage capacity. Full genome sequencing would have been nearly impossible before the advent of the microprocessor, computers, and the Information Age.
A 2015 study[33] done at Children's Mercy Hospital in Kansas City detailed the use of full genome sequencing including full analysis. The process took a record breaking 26 hours[34] and was done using Illumina HiSeq machines, the Edico Genome Dragen Processor, and several custom designed software packages. Most of this acceleration was achieved using the newly developed Dragen Processor which brought the analysis time down from 15 hours to 40 minutes.
Commercialization
A number of public and private companies are competing to develop a full genome sequencing platform that is commercially robust for both research and clinical use,[35] including Illumina,[36] Knome,[37] Sequenom,[38] 454 Life Sciences,[39] Pacific Biosciences,[40] Complete Genomics,[41] Helicos Biosciences,[42] GE Global Research (General Electric), Affymetrix, IBM, Intelligent Bio-Systems,[43] Life Technologies and Oxford Nanopore Technologies.[44] These companies are heavily financed and backed by venture capitalists, hedge funds, and investment banks.[45][46]
Incentive
In October 2006, the X Prize Foundation, working in collaboration with the J. Craig Venter Science Foundation, established the Archon X Prize for Genomics,[47] intending to award US$10 million to "the first Team that can build a device and use it to sequence 100 human genomes within 10 days or less, with an accuracy of no more than one error in every 1,000,000 bases sequenced, with sequences accurately covering at least 98% of the genome, and at a recurring cost of no more than $1,000 per genome".[48] An error rate of 1 in 1,000,000 bases, out of a total of approximately six billion bases in the human diploid genome, would mean about 6,000 errors per genome. The error rates required for widespread clinical use, such as predictive medicine[49] is currently set by over 1,400 clinical single gene sequencing tests[50] (for example, errors in BRCA1 gene for breast cancer risk analysis).
The Archon X Prize for Genomics was cancelled in 2013, before its official start date.[51][52]
History
In 2007, Applied Biosystems started selling a new type of sequencer called SOLiD System.[53] The technology allowed users to sequence 60 gigabases per run.[54]
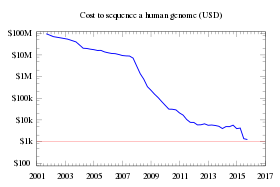
In June 2009, Illumina announced that they were launching their own Personal Full Genome Sequencing Service at a depth of 30× for $48,000 per genome.[55][56]
In August 2009, the founder of Helicos Biosciences, Stephen Quake, stated that using the company's Single Molecule Sequencer he sequenced his own full genome for less than $50,000.[57]
In November 2009, Complete Genomics published a peer-reviewed paper in Science demonstrating its ability to sequence a complete human genome for $1,700.[58][59]
In May 2011, Illumina lowered its Full Genome Sequencing service to $5,000 per human genome, or $4,000 if ordering 50 or more.[60] Helicos Biosciences, Pacific Biosciences, Complete Genomics, Illumina, Sequenom, ION Torrent Systems, Halcyon Molecular, NABsys, IBM, and GE Global appear to all be going head to head in the race to commercialize full genome sequencing.[31][61]
A series of publications in 2012 showed the utility of SMRT sequencing from Pacific Biosciences in generating full genome sequences with de novo assembly.[62]
With sequencing costs declining, a number of companies began claiming that their equipment would soon achieve the $1,000 genome: these companies included Life Technologies in January 2012,[63] Oxford Nanopore Technologies in February 2012[64] and Illumina in February 2014.[65][66]
However, as of 2015, the NHGRI estimates the cost of obtaining a whole-genome sequence at around $1,500.[67]
Comparison with DNA microarrays
Full genome sequencing provides information on a genome that is orders of magnitude larger than that provided by the previous leader in genotyping technology, DNA arrays. For humans, DNA arrays currently provide genotypic information on up to one million genetic variants,[68][69][70] while full genome sequencing will provide information on all six billion bases in the human genome, or 3,000 times more data. Because of this, full genome sequencing is considered a disruptive innovation to the DNA array markets as the accuracy of both range from 99.98% to 99.999% (in non-repetitive DNA regions) and their consumables cost of $5000 per 6 billion base pairs is competitive (for some applications) with DNA arrays ($500 per 1 million basepairs).[39] Agilent, another established DNA array manufacturer, is working on targeted (selective region) genome sequencing technologies.[71] It is thought that Affymetrix, the pioneer of array technology in the 1990s, has fallen behind due to significant corporate and stock turbulence and is currently not working on any known full genome sequencing approach.[72][73][74] It is unknown what will happen to the DNA array market once full genome sequencing becomes commercially widespread, especially as companies and laboratories providing this disruptive technology start to realize economies of scale. It is postulated, however, that this new technology may significantly diminish the total market size for arrays and any other sequencing technology once it becomes commonplace for individuals and newborns to have their full genomes sequenced.[75]
Applications
Mutation frequencies
Whole genome sequencing has established the mutation frequency for whole human genomes. The mutation frequency in the whole genome between generations for humans (parent to child) is about 70 new mutations per generation.[76][77] An even lower level of variation was found comparing whole genome sequencing in blood cells for a pair of monozygotic (identical twins) 100-year-old centenarians.[78] Only 8 somatic differences were found, though somatic variation occurring in less than 20% of blood cells would be undetected.
In the specifically protein coding regions of the human genome, it is estimated that there are about 0.35 mutations that would change the protein sequence between parent/child generations (less than one mutated protein per generation).[79]
In cancer, mutation frequencies are much higher, due to genome instability. This frequency can further depend on patient age, exposure to DNA damaging agents (such as UV-irradiation or components of tobacco smoke) and the activity/inactivity of DNA repair mechanisms.[80] Furthermore, mutation frequency can vary between cancer types: in germline cells, mutation rates occur at approximately 0.023 mutations per megabase, but this number is much higher in breast cancer (1.18-1.66 mutations per Mb), in lung cancer (17.7) or in melanomas (~33).[81]
Diagnostic use and societal impact
Inexpensive, time-efficient full genome sequencing will be a major accomplishment not only for the field of genomics, but for the entire human civilization because, for the first time, individuals will be able to have their entire genome sequenced. Utilizing this information, it is speculated that health care professionals, such as physicians and genetic counselors, will eventually be able to use genomic information to predict what diseases a person may get in the future and attempt to either minimize the impact of that disease or avoid it altogether through the implementation of personalized, preventive medicine. Full genome sequencing will allow health care professionals to analyze the entire human genome of an individual and therefore detect all disease-related genetic variants, regardless of the genetic variant's prevalence or frequency. This will enable the rapidly emerging medical fields of predictive medicine and personalized medicine and will mark a significant leap forward for the clinical genetic revolution. Full genome sequencing is clearly of great importance for research into the basis of genetic disease and has shown significant benefit to a subset of individuals with rare disease in the clinical setting.[82][83][84][85] Illumina's CEO, Jay Flatley, stated in February 2009 that "A complete DNA read-out for every newborn will be technically feasible and affordable in less than five years, promising a revolution in healthcare" and that "by 2019 it will have become routine to map infants' genes when they are born".[86] This potential use of genome sequencing is highly controversial, as it runs counter to established ethical norms for predictive genetic testing of asymptomatic minors that have been well established in the fields of medical genetics and genetic counseling.[87][88][89][90] The traditional guidelines for genetic testing have been developed over the course of several decades since it first became possible to test for genetic markers associated with disease, prior to the advent of cost-effective, comprehensive genetic screening. It is established that norms, such as in the sciences and the field of genetics, are subject to change and evolve over time.[91][92] It is unknown whether traditional norms practiced in medical genetics today will be altered by new technological advancements such as full genome sequencing.
In March 2010, researchers from the Medical College of Wisconsin announced the first successful use of whole-genome sequencing to inform the treatment of a patient.[93][94]
Currently available newborn screening for childhood diseases allows detection of rare disorders that can be prevented or better treated by early detection and intervention. Specific genetic tests are also available to determine an etiology when a child's symptoms appear to have a genetic basis. Full genome sequencing, in addition has the potential to reveal a large amount of information (such as carrier status for autosomal recessive disorders, genetic risk factors for complex adult-onset diseases, and other predictive medical and non-medical information) that is currently not completely understood, may not be clinically useful to the child during childhood, and may not necessarily be wanted by the individual upon reaching adulthood.[95] In addition to predicting disease risk in childhood, genetic testing may have other benefits (such as discovery of non-paternity) but may also have potential downsides (genetic discrimination, loss of anonymity, and psychological impacts).[96] Many publications regarding ethical guidelines for predictive genetic testing of asymptomatic minors may therefore have more to do with protecting minors and preserving the individual's privacy and autonomy to know or not to know their genetic information, than with the technology that makes the tests themselves possible.[97]
Due to recent cost reductions (see above) whole genome sequencing has become a realistic application in DNA diagnostics. In 2013, the 3Gb-TEST consortium obtained funding from the European Union to prepare the health care system for these innovations in DNA diagnostics.[98][99] Quality assessment schemes, Health technology assessment and guidelines have to be in place. The 3Gb-TEST consortium has identified the analysis and interpretation of sequence data as the most complicated step in the diagnostic process.[100] At the Consortium meeting in Athens in September 2014, the Consortium coined the word genotranslation for this crucial step. This step leads to a so-called genoreport. Guidelines are needed to determine the required content of these reports.
Ethical concerns
The majority of ethicists insist that the privacy of individuals undergoing genetic testing must be protected under all circumstances.[101] Data obtained from whole genome sequencing can not only reveal much information about the individual who is the source of DNA, but it can also reveal much probabilistic information about the DNA sequence of close genetic relatives.[102] Furthermore, the data obtained from whole genome sequencing can also reveal much useful predictive information about the relatives present and future health risks.[103] This raises important questions about what obligations, if any, are owed to the family members of the individuals who are undergoing genetic testing. In the Western/European society, tested individuals are usually encouraged to share important information on the genetic diagnosis with their close relatives since the importance of the genetic diagnosis for offspring and other close relatives is usually one of the reasons for seeking a genetic testing in the first place.[101] Nevertheless, a major ethical dilemma can develop when the patients refuse to share information on a diagnosis that is made for serious genetic disorder that is highly preventable and where there is a high risk to relatives carrying the same disease mutation.[102] Under such circumstances, the clinician may suspect that the relatives would rather know of the diagnosis and hence the clinician can face a conflict of interest with respect to patient-doctor confidentiality.[102]
Another major privacy concern is the scientific need to put information on patient's genotypes and phenotypes into the public scientific databases such as the locus specific databases.[102] Although only anonymous patient data are submitted to the locus specific databases, patients might still be identifiable by their relatives in the case of finding a rare disease or a rare missense mutation.[102]
People with public genome sequences
The first nearly complete human genomes sequenced were two Americans of predominantly Northwestern European ancestry in 2007 (J. Craig Venter at 7.5-fold coverage,[104][105][106] and James Watson at 7.4-fold).[107][108][109] This was followed in 2008 by sequencing of an anonymous Han Chinese man (at 36-fold),[110] a Yoruban man from Nigeria (at 30-fold),[111] and a female caucasian Leukemia patient (at 33 and 14-fold coverage for tumor and normal tissues).[112] Steve Jobs was among the first 20 people to have their whole genome sequenced, reportedly for the cost of $100,000.[113] As of June 2012, there are 69 nearly complete human genomes publicly available.[114] Commercialization of full genome sequencing is in an early stage and growing rapidly.
See also
References
- ↑ Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2008). "Chapter 8". Molecular biology of the cell (5th ed.). New York: Garland Science. p. 550. ISBN 0-8153-4106-7.
- ↑ Kijk magazine, 01 January 2009
- ↑ Gilissen (Jul 2014). "Genome sequencing identifies major causes of severe intellectual disability". Nature. 511 (7509): 344–7. doi:10.1038/nature13394. PMID 24896178.
- ↑ Nones, K; Waddell, N; Wayte, N; Patch, AM; Bailey, P; Newell, F; Holmes, O; Fink, JL; Quinn, MC; Tang, YH; Lampe, G; Quek, K; Loffler, KA; Manning, S; Idrisoglu, S; Miller, D; Xu, Q; Waddell, N; Wilson, PJ; Bruxner, TJ; Christ, AN; Harliwong, I; Nourse, C; Nourbakhsh, E; Anderson, M; Kazakoff, S; Leonard, C; Wood, S; Simpson, PT; Reid, LE; Krause, L; Hussey, DJ; Watson, DI; Lord, RV; Nancarrow, D; Phillips, WA; Gotley, D; Smithers, BM; Whiteman, DC; Hayward, NK; Campbell, PJ; Pearson, JV; Grimmond, SM; Barbour, AP (29 October 2014). "Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis". Nature Communications. 5: 5224. doi:10.1038/ncomms6224. PMID 25351503.
- ↑ van El, CG; Cornel, MC; Borry, P; Hastings, RJ; Fellmann, F; Hodgson, SV; Howard, HC; Cambon-Thomsen, A; Knoppers, BM; Meijers-Heijboer, H; Scheffer, H; Tranebjaerg, L; Dondorp, W; de Wert, GM (June 2013). "Whole-genome sequencing in health care. Recommendations of the European Society of Human Genetics". European Journal of Human Genetics. 21 Suppl 1: S1–5. doi:10.1038/ejhg.2013.46. PMID 23819146.
- ↑ Mooney, Sean (Sep 2014). "Progress towards the integration of pharmacogenomics in practice". Human Genetics. doi:10.1007/s00439-014-1484-7. PMID 25238897.
- ↑ Marx, Vivien (11 September 2013). "Next-generation sequencing: The genome jigsaw". Nature. 501 (7466): 263–268. doi:10.1038/501261a.
- ↑ al.], Bruce Alberts ... [et (2008). Molecular biology of the cell (5th ed.). New York: Garland Science. p. 551. ISBN 0-8153-4106-7.
- 1 2 3 Fleischmann, R.; Adams, M.; White, O; Clayton, R.; Kirkness, E.; Kerlavage, A.; Bult, C.; Tomb, J.; Dougherty, B.; Merrick, J.; al., e. (28 July 1995). "Whole-genome random sequencing and assembly of Haemophilus influenzae Rd". Science. 269 (5223): 496–512. doi:10.1126/science.7542800. PMID 7542800.
- ↑ Eddy, Sean R. (November 2012). "The C-value paradox, junk DNA and ENCODE". Current Biology. 22 (21): R898–R899. doi:10.1016/j.cub.2012.10.002. PMID 23137679.
- ↑ PELLICER, JAUME; FAY, MICHAEL F.; LEITCH, ILIA J. (15 September 2010). "The largest eukaryotic genome of them all?". Botanical Journal of the Linnean Society. 164 (1): 10–15. doi:10.1111/j.1095-8339.2010.01072.x.
- ↑ Human Genome Sequencing Consortium, International (21 October 2004). "Finishing the euchromatic sequence of the human genome". Nature. 431 (7011): 931–945. doi:10.1038/nature03001. PMID 15496913.
- ↑ Goffeau, A.; Barrell, B. G.; Bussey, H.; Davis, R. W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J. D.; Jacq, C.; Johnston, M.; Louis, E. J.; Mewes, H. W.; Murakami, Y.; Philippsen, P.; Tettelin, H.; Oliver, S. G. (25 October 1996). "Life with 6000 Genes" (PDF). Science. 274 (5287): 546–567. doi:10.1126/science.274.5287.546. PMID 8849441.
- ↑ The C. elegans Sequencing Consortium (11 December 1998). "Genome Sequence of the Nematode C. elegans: A Platform for Investigating Biology". Science. 282 (5396): 2012–2018. doi:10.1126/science.282.5396.2012. PMID 9851916.
- ↑ Alberts, Bruce (2008). Molecular Biology of the Cell (5th ed.). New York: Garland Science. p. 552. ISBN 0-8153-4106-7.
- ↑ Dunham, I. "The DNA sequence of human chromosome 22". nature.com.
- ↑ Adams MD; Celniker SE; Holt RA; et al. (2000-03-24). "The Genome Sequence of Drosophila melanogaster". Science. 287: 2185–2195. doi:10.1126/science.287.5461.2185. PMID 10731132.
- ↑ "Analysis of the genome sequence of the flowering plant Arabidopsis thaliana.". Nature. 408: 796–815. 2000-12-14. doi:10.1038/35048692. PMID 11130711.
- ↑ Venter JC; Adams MD; Myers EW; et al. (2001-02-16). "The Sequence of the Human Genome". Science. 291: 1304–1351. doi:10.1126/science.1058040. PMID 11181995.
- ↑ Waterston RH; Lindblad-Toh K; Birney E; et al. (2002-10-31). "Initial sequencing and comparative analysis of the mouse genome". Nature. 420: 520–562. doi:10.1038/nature01262. PMID 12466850.
- ↑ "Finishing the euchromatic sequence of the human genome". Nature. 431: 931–945. 2004-09-07. doi:10.1038/nature03001. PMID 15496913.
- ↑ Braslavsky, Ido; et al. (2003). "Sequence information can be obtained from single DNA molecules". Proc Natl Acad Sci USA. 100 (7): 3960–3984. doi:10.1073/pnas.0230489100. PMC 153030
 . PMID 12651960.
. PMID 12651960. - ↑ Single-cell Sequencing Makes Strides in the Clinic with Cancer and PGD First Applications from Clinical Sequencing News. By Monica Heger. October 02, 2013
- ↑ Yurkiewicz, I. R.; Korf, B. R.; Lehmann, L. S. (2014). "Prenatal whole-genome sequencing--is the quest to know a fetus's future ethical?". New England Journal of Medicine. 370 (3): 195–7. doi:10.1056/NEJMp1215536. PMID 24428465.
- ↑ Staden R (June 1979). "A strategy of DNA sequencing employing computer programs". Nucleic Acids Res. 6 (7): 2601–10. doi:10.1093/nar/6.7.2601. PMC 327874
 . PMID 461197.
. PMID 461197. - ↑ Edwards, A; Caskey, T (1991). "Closure strategies for random DNA sequencing". Methods: A Companion to Methods in Enzymology. 3 (1): 41–47. doi:10.1016/S1046-2023(05)80162-8.
- ↑ Edwards A; Voss H; Rice P; Civitello A; Stegemann J; Schwager C; Zimmermann J; Erfle H; Caskey CT; Ansorge W (April 1990). "Automated DNA sequencing of the human HPRT locus". Genomics. 6 (4): 593–608. doi:10.1016/0888-7543(90)90493-E. PMID 2341149.
- ↑ Roach JC; Boysen C; Wang K; Hood L (March 1995). "Pairwise end sequencing: a unified approach to genomic mapping and sequencing". Genomics. 26 (2): 345–53. doi:10.1016/0888-7543(95)80219-C. PMID 7601461.
- ↑ Fleischmann RD; Adams MD; White O; Clayton RA; Kirkness EF; Kerlavage AR; Bult CJ; Tomb JF; Dougherty BA; Merrick JM; McKenney; Sutton; Fitzhugh; Fields; Gocyne; Scott; Shirley; Liu; Glodek; Kelley; Weidman; Phillips; Spriggs; Hedblom; Cotton; Utterback; Hanna; Nguyen; Saudek; et al. (July 1995). "Whole-genome random sequencing and assembly of Haemophilus influenzae Rd". Science. 269 (5223): 496–512. Bibcode:1995Sci...269..496F. doi:10.1126/science.7542800. PMID 7542800.
- ↑ Adams, MD; et al. (2000). "The genome sequence of Drosophila melanogaster". Science. 287 (5461): 2185–95. Bibcode:2000Sci...287.2185.. doi:10.1126/science.287.5461.2185. PMID 10731132.
- 1 2 Mukhopadhyay R (February 2009). "DNA sequencers: the next generation". Anal. Chem. 81 (5): 1736–40. doi:10.1021/ac802712u. PMID 19193124.
- ↑ Kwong, JC; McCallum, N; Sintchenko, V; Howden, BP (April 2015). "Whole genome sequencing in clinical and public health microbiology.". Pathology. 47 (3): 199–210. PMC 4389090
 . PMID 25730631.
. PMID 25730631. - ↑ "A 26-hour system of highly sensitive whole genome sequencing for emergency management of genetic diseases | Genome Medicine | Full Text". Genome Medicine. 2015-07-13. Retrieved 2016-11-11.
- ↑ Strickland, Eliza (2015-10-14). "New Genetic Technologies Diagnose Critically Ill Infants Within 26 Hours - IEEE Spectrum". Spectrum.ieee.org. Retrieved 2016-11-11.
- ↑ "Article : Race to Cut Whole Genome Sequencing Costs Genetic Engineering & Biotechnology News — Biotechnology from Bench to Business". Genengnews.com. Retrieved 2009-02-23.
- ↑ "Whole Genome Sequencing Costs Continue to Drop". Eyeondna.com. Retrieved 2009-02-23.
- ↑ Harmon, Katherine (2010-06-28). "Genome Sequencing for the Rest of Us". Scientific American. Retrieved 2010-08-13.
- ↑ San Diego/Orange County Technology News. "Sequenom to Develop Third-Generation Nanopore-Based Single Molecule Sequencing Technology". Freshnews.com. Retrieved 2009-02-24.
- 1 2 "Article : Whole Genome Sequencing in 24 Hours Genetic Engineering & Biotechnology News — Biotechnology from Bench to Business". Genengnews.com. Retrieved 2009-02-23.
- ↑ "Pacific Bio lifts the veil on its high-speed genome-sequencing effort". VentureBeat. Retrieved 2009-02-23.
- ↑ "Bio-IT World". Bio-IT World. 2008-10-06. Retrieved 2009-02-23.
- ↑ "With New Machine, Helicos Brings Personal Genome Sequencing A Step Closer". Xconomy. 2008-04-22. Retrieved 2011-01-28.
- ↑ "Whole genome sequencing costs continue to fall: $300 million in 2003, $1 million 2007, $60,000 now, $5000 by year end". Nextbigfuture.com. 2008-03-25. Retrieved 2011-01-28.
- ↑ "Han Cao's nanofluidic chip could cut DNA sequencing costs dramatically". Technology Review.
- ↑ John Carroll (2008-07-14). "Pacific Biosciences gains $100M for sequencing tech". FierceBiotech. Retrieved 2009-02-23.
- ↑ Sibley, Lisa (2009-02-08). "Complete Genomics brings radical reduction in cost". Silicon Valley / San Jose Business Journal. Sanjose.bizjournals.com. Retrieved 2009-02-23.
- ↑ Carlson, Rob (2007-01-02). "A Few Thoughts on Rapid Genome Sequencing and The Archon Prize — synthesis". Synthesis.cc. Retrieved 2009-02-23.
- ↑ "PRIZE Overview: Archon X PRIZE for Genomics". Archived December 2, 2008, at the Wayback Machine.
- ↑ Bentley DR (December 2006). "Whole-genome re-sequencing". Curr. Opin. Genet. Dev. 16 (6): 545–552. doi:10.1016/j.gde.2006.10.009. PMID 17055251.
- ↑ "Home". GeneTests. Retrieved 2016-11-11.
- ↑ Diamandis, Peter. "Outpaced by Innovation: Canceling an XPRIZE". Huffington Post.
- ↑ Aldhous, Peter. "X Prize for genomes cancelled before it begins".
- ↑ "SOLiD System — a next-gen DNA sequencing platform announced". Gizmag.com. 2007-10-27. Retrieved 2009-02-24.
- ↑ "The $1000 Genome: Coming Soon?". Dddmag.com. 2010-04-01. Retrieved 2011-01-28.
- ↑ "Individual genome sequencing — Illumina, Inc.". Everygenome.com. Retrieved 2011-01-28.
- ↑ "Illumina launches personal genome sequencing service for $48,000 : Genetic Future". Scienceblogs.com. Archived from the original on June 16, 2009. Retrieved 2011-01-28.
- ↑ Wade, Nicholas (2009-08-11). "Cost of Decoding a Genome Is Lowered". The New York Times. Retrieved 2010-05-03.
- ↑
- ↑ Drmanac R, Sparks AB, Callow MJ et al.: Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays" Science 327(5961), 78-81 (2010)
- ↑
- ↑ "NHGRI Awards More than $50M for Low-Cost DNA Sequencing Tech Development". Genome Web. 2009.
- ↑ "Finished bacterial genomes from shotgun sequence data" (PDF).
- ↑ "Life Technologies Introduces the Benchtop Ion Proton™ Sequencer; Designed to Decode a Human Genome in One Day for $1,000" (press release). Archived from the original on December 23, 2012. Retrieved August 30, 2012.
- ↑ ANDREW POLLACK (2012-02-17). "Oxford Nanopore Unveils Tiny DNA Sequencing Device - The New York Times". Nytimes.com. Retrieved 2016-11-11.
- ↑ "Illumina Sequencer Enables $1,000 Genome". News: Genomics & Proteomics. Gen. Eng. Biotechnol. News (paper). 34 (4). 15 February 2014. p. 18.
- ↑ Check Hayden, Erika (15 January 2014). "Is the $1,000 genome for real?". Nature. doi:10.1038/nature.2014.14530.
- ↑ "The Cost of Sequencing a Human Genome". www.genome.gov.
- ↑ "Genomics Core". Gladstone.ucsf.edu. Archived from the original on June 30, 2010. Retrieved 2009-02-23.
- ↑ Nishida N; Koike A; Tajima A; Ogasawara Y; Ishibashi Y; Uehara Y; Inoue I; Tokunaga K (2008). "Evaluating the performance of Affymetrix SNP Array 6.0 platform with 400 Japanese individuals". BMC Genomics. 9 (1): 431. doi:10.1186/1471-2164-9-431. PMC 2566316
 . PMID 18803882.
. PMID 18803882. 
- ↑ Petrone, Justin. "Illumina, DeCode Build 1M SNP Chip; Q2 Launch to Coincide with Release of Affy's 6.0 SNP Array | BioArray News | Arrays". GenomeWeb. Retrieved 2009-02-23.
- ↑ "Agilent Technologies Announces Licensing Agreement with Broad Institute to Develop Genome-Partitioning Kits to Streamline Next-Generation Sequencing". Archived from the original on December 25, 2008.
- ↑ "Affymetrix stock slumps 30% on forecast". Sacramento Business Journal. Sacramento.bizjournals.com. 2008-07-25. Retrieved 2009-02-23.
- ↑ Bluis, John (2006-04-24). "Affymetrix Gets Chipped Again". Fool.com. Retrieved 2009-02-23.
- ↑ "The chips are down". Nature. 444 (7117): 256–7. November 2006. Bibcode:2006Natur.444..256.. doi:10.1038/444256a. PMID 17108930.
- ↑ Coombs A (October 2008). "The sequencing shakeup". Nat. Biotechnol. 26 (10): 1109–12. doi:10.1038/nbt1008-1109. PMID 18846083.
- ↑ Roach JC; Glusman G; Smit AF; et al. (April 2010). "Analysis of genetic inheritance in a family quartet by whole-genome sequencing". Science. 328 (5978): 636–9. doi:10.1126/science.1186802. PMC 3037280
 . PMID 20220176.
. PMID 20220176. - ↑ Campbell CD; Chong JX; Malig M; et al. (November 2012). "Estimating the human mutation rate using autozygosity in a founder population". Nat. Genet. 44 (11): 1277–81. doi:10.1038/ng.2418. PMC 3483378
 . PMID 23001126.
. PMID 23001126. - ↑ Ye K; Beekman M; Lameijer EW; Zhang Y; Moed MH; van den Akker EB; Deelen J; Houwing-Duistermaat JJ; Kremer D; Anvar SY; Laros JF; Jones D; Raine K; Blackburne B; Potluri S; Long Q; Guryev V; van der Breggen R; Westendorp RG; 't Hoen PA; den Dunnen J; van Ommen GJ; Willemsen G; Pitts SJ; Cox DR; Ning Z; Boomsma DI; Slagboom PE (December 2013). "Aging as accelerated accumulation of somatic variants: whole-genome sequencing of centenarian and middle-aged monozygotic twin pairs". Twin Res Hum Genet. 16 (6): 1026–32. doi:10.1017/thg.2013.73. PMID 24182360.
- ↑ Keightley PD (February 2012). "Rates and fitness consequences of new mutations in humans". Genetics. 190 (2): 295–304. doi:10.1534/genetics.111.134668. PMC 3276617
 . PMID 22345605.
. PMID 22345605. - ↑ Milholland B; Auton A; Suh Y; Vijg J (September 22, 2015). "Age-related somatic mutations in the cancer genome". Oncotarget. 6 (28): 24627–35. doi:10.18632/oncotarget.5685. PMC 4694783
 . PMID 26384365.
. PMID 26384365. - ↑ Tuna M; Amos CI (November 2013). "Genomic sequencing in cancer". Cancer Lett. 340 (2): 161–70. doi:10.1016/j.canlet.2012.11.004. PMID 23178448.
- ↑ Ng SB; Buckingham KJ; Lee C; et al. (January 2010). "Exome sequencing identifies the cause of a mendelian disorder". Nat. Genet. 42 (1): 30–5. doi:10.1038/ng.499. PMC 2847889
 . PMID 19915526.
. PMID 19915526. - ↑ Hannibal MC; Buckingham KJ; Ng SB; et al. (July 2011). "Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome". Am. J. Med. Genet. A. 155A (7): 1511–6. doi:10.1002/ajmg.a.34074. PMC 3121928
 . PMID 21671394.
. PMID 21671394. - ↑ Worthey EA; Mayer AN; Syverson GD; et al. (March 2011). "Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease". Genet. Med. 13 (3): 255–62. doi:10.1097/GIM.0b013e3182088158. PMID 21173700.
- ↑ Goh V; Helbling D; Biank V; Jarzembowski J; Dimmock D (June 2011). "Next Generation Sequencing Facilitates The Diagnosis In A Child With Twinkle Mutations Causing Cholestatic Liver Failure". J Pediatr Gastroenterol Nutr. 54 (2): 291–4. doi:10.1097/MPG.0b013e318227e53c. PMID 21681116.
- ↑ Henderson, Mark (2009-02-09). "Genetic mapping of babies by 2019 will transform preventive medicine". London: Times Online. Retrieved 2009-02-23.
- ↑ McCabe LL; McCabe ER (June 2001). "Postgenomic medicine. Presymptomatic testing for prediction and prevention". Clin Perinatol. 28 (2): 425–34. doi:10.1016/S0095-5108(05)70094-4. PMID 11499063.
- ↑ Nelson RM; Botkjin JR; Kodish ED; et al. (June 2001). "Ethical issues with genetic testing in pediatrics". Pediatrics. 107 (6): 1451–5. doi:10.1542/peds.107.6.1451. PMID 11389275.
- ↑ Borry P; Fryns JP; Schotsmans P; Dierickx K (February 2006). "Carrier testing in minors: a systematic review of guidelines and position papers". Eur. J. Hum. Genet. 14 (2): 133–8. doi:10.1038/sj.ejhg.5201509. PMID 16267502.
- ↑ Borry P; Stultiens L; Nys H; Cassiman JJ; Dierickx K (November 2006). "Presymptomatic and predictive genetic testing in minors: a systematic review of guidelines and position papers". Clin. Genet. 70 (5): 374–81. doi:10.1111/j.1399-0004.2006.00692.x. PMID 17026616.
- ↑ Mesoudi A; Danielson P (August 2008). "Ethics, evolution and culture". Theory Biosci. 127 (3): 229–40. doi:10.1007/s12064-008-0027-y. PMID 18357481.
- ↑ Ehrlich PR; Levin SA (June 2005). "The evolution of norms". PLoS Biol. 3 (6): e194. doi:10.1371/journal.pbio.0030194. PMC 1149491
 . PMID 15941355.
. PMID 15941355. 
- ↑ "One In A Billion: A boy's life, a medical mystery". Jsonline.com. Retrieved 2016-11-11.
- ↑ Abbott, Phil. "US clinics quietly embrace whole-genome sequencing : Nature News". Nature.com. Retrieved 2016-11-11.
- ↑ Mayer AN; Dimmock DP; Arca MJ; et al. (March 2011). "A timely arrival for genomic medicine". Genet. Med. 13 (3): 195–6. doi:10.1097/GIM.0b013e3182095089. PMID 21169843.
- ↑ Ayday E; De Cristofaro E; Hubaux JP; Tsudik G (2015). "The Chills and Thrills of Whole Genome Sequencing". arXiv:1306.1264
 .
. - ↑ Borry, P.; Evers-Kiebooms, G.; Cornel, MC; Clarke, A; Dierickx, K; Public Professional Policy Committee (PPPC) of the European Society of Human Genetics (ESHG) (2009). "Genetic testing in asymptomatic minors Background considerations towards ESHG Recommendations". Eur J Hum Genet. 17 (6): 711–9. doi:10.1038/ejhg.2009.25. PMC 2947094
 . PMID 19277061.
. PMID 19277061. - ↑ "Introducing diagnostic applications of '3Gb-testing' in human genetics".
- ↑ "Beyond public health genomics: proposals from an international working group". Eur J Public Health. 24: 877–879. Aug 2014. doi:10.1093/eurpub/cku142. PMID 25168910.
- ↑ "RD-Connect News 18 July 2014". Rd-connect.eu. Retrieved 2016-11-11.
- 1 2 Sijmons, R.H; Van Langen, I.M (2011). "A clinical perspective on ethical issues in genetic issues". Accountability in Research: Policies and Quality Assurance. 18 (3): 148–162. doi:10.1080/08989621.2011.575033.
- 1 2 3 4 5 Sijmons, R.H.; Van Langen, I.M (2011). "A clinical perspective on ethical issues in genetic testing". Accountability in Research: Policies and Quality Assurance. 18 (3): 148–162. doi:10.1080/08989621.2011.575033.
- ↑ McGuire, Amy, L; Caulfield, Timothy (2008). "Science and Society: Research ethics and the challenge of whole-genome sequencing". Nature Reviews Genetics. 9 (2): 152–156. doi:10.1038/nrg2302.
- ↑ Wade, Nicholas (September 4, 2007). "In the Genome Race, the Sequel Is Personal". New York Times. Retrieved February 22, 2009.
- ↑ Nature. "Access : All about Craig: the first 'full' genome sequence". Nature. Retrieved 2009-02-24.
- ↑ Levy S; Sutton G; Ng PC; Feuk L; Halpern AL; Walenz BP; Axelrod N; Huang J; Kirkness EF; Denisov G; Lin Y; MacDonald JR; Pang AW; Shago M; Stockwell TB; Tsiamouri A; Bafna V; Bansal V; Kravitz SA; Busam DA; Beeson KY; McIntosh TC; Remington KA; Abril JF; Gill J; Borman J; Rogers YH; Frazier ME; Scherer SW; Strausberg RL; Venter JC (September 2007). "The diploid genome sequence of an individual human". PLoS Biol. 5 (10): e254. doi:10.1371/journal.pbio.0050254. PMC 1964779
 . PMID 17803354.
. PMID 17803354. 
- ↑ Wade, Wade (June 1, 2007). "DNA pioneer Watson gets own genome map". International Herald Tribune. Archived from the original on September 27, 2008. Retrieved February 22, 2009.
- ↑ Wade, Nicholas (May 31, 2007). "Genome of DNA Pioneer Is Deciphered". New York Times. Retrieved February 21, 2009.
- ↑ Wheeler DA; Srinivasan M; Egholm M; Shen Y; Chen L; McGuire A; He W; Chen YJ; Makhijani V; Roth GT; Gomes X; Tartaro K; Niazi F; Turcotte CL; Irzyk GP; Lupski JR; Chinault C; Song XZ; Liu Y; Yuan Y; Nazareth L; Qin X; Muzny DM; Margulies M; Weinstock GM; Gibbs RA; Rothberg JM (2008). "The complete genome of an individual by massively parallel DNA sequencing". Nature. 452 (7189): 872–6. Bibcode:2008Natur.452..872W. doi:10.1038/nature06884. PMID 18421352.
- ↑ Wang J; Wang, Wei; Li, Ruiqiang; Li, Yingrui; Tian, Geng; Goodman, Laurie; Fan, Wei; Zhang, Junqing; Li, Jun; Zhang, Juanbin, Juanbin; Guo, Yiran, Yiran; Feng, Binxiao, Binxiao; Li, Heng, Heng; Lu, Yao, Yao; Fang, Xiaodong, Xiaodong; Liang, Huiqing, Huiqing; Du, Zhenglin, Zhenglin; Li, Dong, Dong; Zhao, Yiqing, Yiqing; Hu, Yujie, Yujie; Yang, Zhenzhen, Zhenzhen; Zheng, Hancheng, Hancheng; Hellmann, Ines, Ines; Inouye, Michael, Michael; Pool, John, John; Yi, Xin, Xin; Zhao, Jing, Jing; Duan, Jinjie, Jinjie; Zhou, Yan, Yan; et al. (2008). "The diploid genome sequence of an Asian individual". Nature. 456 (7218): 60–65. Bibcode:2008Natur.456...60W. doi:10.1038/nature07484. PMC 2716080
 . PMID 18987735.
. PMID 18987735. - ↑ Bentley DR; Balasubramanian S; et al. (2008). "Accurate whole human genome sequencing using reversible terminator chemistry". Nature. 456 (7218): 53–9. Bibcode:2008Natur.456...53B. doi:10.1038/nature07517. PMC 2581791
 . PMID 18987734.
. PMID 18987734. - ↑ Ley TJ; Mardis ER; Ding L; Fulton B; McLellan MD; Chen K; Dooling D; Dunford-Shore BH; McGrath S; Hickenbotham M; Cook L; Abbott R; Larson DE; Koboldt DC; Pohl C; Smith S; Hawkins A; Abbott S; Locke D; Hillier LW; Miner T; Fulton L; Magrini V; Wylie T; Glasscock J; Conyers J; Sander N; Shi X; Osborne JR; et al. (2008). "DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome". Nature. 456 (7218): 66–72. Bibcode:2008Natur.456...66L. doi:10.1038/nature07485. PMC 2603574
 . PMID 18987736.
. PMID 18987736. - ↑ Lohr, Steve (2011-10-20). "New Book Details Jobs's Fight Against Cancer". The New York Times.
- ↑ "Complete Human Genome Sequencing Datasets to its Public Genomic Repository". Archived from the original on June 10, 2012.
External links
- Archon X Prize for Genomics
- James Watson's Personal Genome Sequence
- AAAS/Science: Genome Sequencing Poster
- Outsmart Your Genes: Book that discusses full genome sequencing and its impact upon health care and society
- Whole genome linkage analysis