Separase
| Separase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 3.4.22.49 | ||||||||
| CAS number | 351527-77-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
| View/Edit Human | View/Edit Mouse |
Separase, also known as separin, is a cysteine protease responsible for triggering anaphase by hydrolysing cohesin, which is the protein responsible for binding sister chromatids during the early stage of anaphase.[3] In humans, separin is encoded by the ESPL1 gene.[4]
Discovery
In S. cerevisiae, separase is encoded by the esp1 gene. Esp1 was discovered by Kim Nasmyth and coworkers in 1998.[5][6]
Function
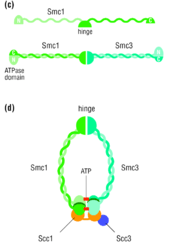
Stable cohesion between sister chromatids before anaphase and their timely separation during anaphase are critical for cell division and chromosome inheritance. In vertebrates, sister chromatid cohesion is released in 2 steps via distinct mechanisms. The first step involves phosphorylation of STAG1 or STAG2 in the cohesin complex. The second step involves cleavage of the cohesin subunit SCC1 (RAD21) by separase, which initiates the final separation of sister chromatids.[8]
In S. cerevisiae, Esp1 is coded by ESP1 and is regulated by the securin Pds1. The two sister chromatids are initially bound together by the cohesin complex until the beginning of anaphase, at which point the mitotic spindle pulls the two sister chromatids apart, leaving each of the two daughter cells with an equivalent number of sister chromatids. The proteins that bind the two sister chromatids, disallowing any premature sister chromatid separation, are a part of the cohesin protein family. One of these cohesin proteins crucial for sister chromatid cohesion is Scc1. Esp1 is a separase protein that cleaves the cohesin subunit Scc1 (RAD21), allowing sister chromatids to separate at the onset of anaphase during mitosis.[6]
Regulation

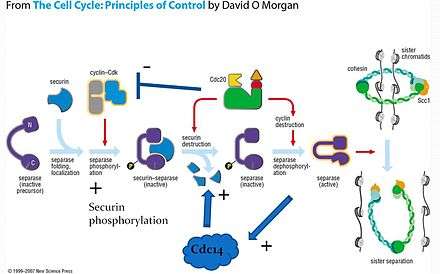
When the cell is not dividing, separase is prevented from cleaving cohesin through its association with another protein, securin, as well as phosphorylation by the cyclin-CDK complex. This provides two layers of negative regulation to prevent inappropriate cohesin cleavage. Note that separase cannot function without initially forming the securin-separase complex in most organisms. This is because securin helps properly fold separase into the functional conformation. However, yeast does not appear to require securin to form functional separase because anaphase occurs in yeast even with a securin deletion.[7]
On the signal for anaphase, securin is ubiquitinated and hydrolysed, releasing separase for dephosphorylation by the APC-Cdc20 complex. Active separase can then cleave Scc1 for release of the sister chromatids.
Separase initiates the activation of Cdc14 in early anaphase[10] and Cdc14 has been found to dephosphorylate securin, thereby increasing its efficiency as a substrate for degradation. The presence of this positive feedback loop offers a potential mechanism for giving anaphase a more switch-like behavior.[9]
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ "ESPL1 - Separin - Homo sapiens (Human) - ESPL1 gene & protein". Uniprot.org. 2010-10-05. Retrieved 2016-05-14.
- ↑ Nagase T, Seki N, Ishikawa K, Tanaka A, Nomura N (February 1996). "Prediction of the coding sequences of unidentified human genes. V. The coding sequences of 40 new genes (KIAA0161-KIAA0200) deduced by analysis of cDNA clones from human cell line KG-1". DNA Res. 3 (1): 17–24. doi:10.1093/dnares/3.1.17. PMID 8724849.
- ↑ Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K (June 1998). "An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast". Cell. 93 (6): 1067–76. doi:10.1016/S0092-8674(00)81211-8. PMID 9635435.
- 1 2 Uhlmann F; Lottspeich F; Nasmyth K (July 1999). "Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1". Nature. 400 (6739): 37–42. doi:10.1038/21831. PMID 10403247.
- 1 2 Morgan, David O (2007). The cell cycle: principles of control. London: Published by New Science Press in association with Oxford University Press. ISBN 0-87893-508-8.
- ↑ Sun Y, Kucej M, Fan HY, Yu H, Sun QY, Zou H (April 2009). "Separase is recruited to mitotic chromosomes to dissolve sister chromatid cohesion in a DNA-dependent manner". Cell. 137 (1): 123–32. doi:10.1016/j.cell.2009.01.040. PMC 2673135
 . PMID 19345191.
. PMID 19345191. - 1 2 Holt LJ, Krutchinsky AN, Morgan DO (July 2008). "Positive feedback sharpens the anaphase switch". Nature. 454 (7202): 353–7. doi:10.1038/nature07050. PMC 2636747
 . PMID 18552837.
. PMID 18552837. - ↑ Stegmeier F, Visintin R, Amon A (January 2002). "Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase". Cell. 108 (2): 207–20. doi:10.1016/S0092-8674(02)00618-9. PMID 11832211.
Further reading
- McGrew J, Goetsch L, Byers B, Baum P (1992). "Requirement for ESP1 in the nuclear division of Saccharomyces cerevisiae". Mol. Biol. Cell. 3 (12): 1443–54. doi:10.1091/mbc.3.12.1443. PMC 275712
 . PMID 1493337.
. PMID 1493337. - Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K (1998). "An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast". Cell. 93 (6): 1067–76. doi:10.1016/S0092-8674(00)81211-8. PMID 9635435.
- Jensen S, Segal M, Clarke D, Reed S (2001). "A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1". J. Cell Biol. 152 (1): 27–40. doi:10.1083/jcb.152.1.27. PMC 2193664
 . PMID 11149918.
. PMID 11149918.
External links
- separase at the US National Library of Medicine Medical Subject Headings (MeSH)
- http://www.ncbi.nih.gov/entrez/query.fcgi?db=nucleotide&cmd=search&term=L07289&doptcmdl=GenBank
- Video by David Morgan explaining action of securin and separin (in MP4 format): http://media.hhmi.org/ibio/morgan/morgan_3.mp4
- and in other formats: http://www.ibioseminars.org/lectures/bio-mechanisms/david-morgan-part-1/david-morgan-part-3.html
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
