Scaffold protein
In biology, scaffold proteins are crucial regulators of many key signaling pathways. Although scaffolds are not strictly defined in function, they are known to interact and/or bind with multiple members of a signaling pathway, tethering them into complexes. In such pathways, they regulate signal transduction and help localize pathway components (organized in complexes) to specific areas of the cell such as the plasma membrane, the cytoplasm, the nucleus, the Golgi, endosomes, and the mitochondria.
History
The first signaling scaffold protein discovered was the Ste5 protein from the yeast Saccharomyces cerevisiae. Three distinct domains of Ste5 were shown to associate with the protein kinases Ste11, Ste7, and Fus3 to form a multikinase complex.[2]
Function
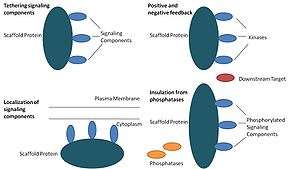
Scaffold proteins act in at least four ways: tethering signaling components, localizing these components to specific areas of the cell, regulating signal transduction by coordinating positive and negative feedback signals, and insulating correct signaling proteins from competing proteins.[1]
Tethering signaling components
This particular function is considered a scaffold's most basic function. Scaffolds assemble signaling components of a cascade into complexes. This assembly may be able to enhance signaling specificity by preventing unnecessary interactions between signaling proteins, and enhance signaling efficiency by increasing the proximity and effective concentration of components in the scaffold complex. A common example of how scaffolds enhance specificity is a scaffold that binds a protein kinase and its substrate, thereby ensuring specific kinase phosphorylation. Additionally, some signaling proteins require multiple interactions for activation and scaffold tethering may be able to convert these interactions into one interaction that results in multiple modifications.[3][4] Scaffolds may also be catalytic as interaction with signaling proteins may result in allosteric changes of these signaling components.[5] Such changes may be able to enhance or inhibit the activation of these signaling proteins. An example is the Ste5 scaffold in the mitogen-activated protein kinase (MAPK) pathway. Ste5 has been proposed to direct mating signaling through the Fus3 MAPK by catalytically unlocking this particular kinase for activation by its MAPKK Ste7.[6]
Localization of signaling components in the cell
Scaffolds localize the signaling reaction to a specific area in the cell, a process that could be important for the local production of signaling intermediates. A particular example of this process is the scaffold, A-kinase anchor proteins (AKAPs), which target cyclic AMP-dependent protein kinase (PKA) to various sites in the cell.[7] This localization is able to locally regulate PKA and results in the local phosphorylation by PKA of its substrates.
Coordinating positive and negative feedback
Many hypotheses about how scaffolds coordinate positive and negative feedback come from engineered scaffolds and mathematical modeling. In three-kinase signaling cascades, scaffolds bind all three kinases, enhancing kinase specificity and restricting signal amplification by limiting kinase phosphorylation to only one downstream target.[3][8][9] These abilities may be related to stability of the interaction between the scaffold and the kinases, the basal phosphatase activity in the cell, scaffold location, and expression levels of the signaling components.[3][8]
Insulating correct signaling proteins from inactivation
Signaling pathways are often inactivated by enzymes that reverse the activation state and/or induce the degradation of signaling components. Scaffolds have been proposed to protect activated signaling molecules from inactivation and/or degradation. Mathematical modeling has shown that kinases in a cascade without scaffolds have a higher probability of being dephosphorylated by phosphatases before they are even able to phosphorylate downstream targets.[8] Furthermore, scaffolds have been shown to insulate kinases from substrate- and ATP-competitive inhibitors.[10]
Scaffold protein summary
| Scaffold Proteins | Pathway | Potential Functions | Description |
|---|---|---|---|
| KSR | MAPK | Assembly and localization of the RAS-ERK pathway | One of the best studied signaling pathways in biology is the RAS-ERK pathway in which the RAS G-protein activates the MAPKKK RAF, which activates the MAPKK MEK1 (MAPK/ERK kinase 1), which then activates the MAPK ERK. Several scaffold proteins have been identified to be involved in this pathway and other similar MAPK pathways. One such scaffold protein is KSR, which is the most probable equivalent of the well-studied yeast MAPK scaffold protein Ste5.[11] It is a positive regulator of the pathway and binds many proteins in the pathway, including all three kinases in the cascade.[6] KSR has been shown to be localized to the plasma membrane during cell activation, thereby playing a role in assembling the components of the ERK pathway and in localizing activated ERK to the plasma membrane.[12] |
| MEKK1 | MAPK | Assembly and localization of the death receptor signalosome | Other scaffold proteins include B-cell lymphoma 10 (BCL-10) and MEK kinase 1 (MEKK1), which have roles in the JUN N-terminal kinase (JNK) pathway. |
| BCL-10 | MAPK | Assembly and specificity of JNK | |
| AKAP | PKA Pathways | Coordination of phosphorylation by PKA onto downstream targets | This family of proteins is only structurally related in their ability to bind the regulatory subunit of PKA but can otherwise bind a very diverse set of enzymes and substrates |
| AHNAK-1 | Calcium signaling | Assembly and localization of calcium channels | Calcium signaling is essential for the proper function of immune cells. Recent studies have shown that the scaffold protein, AHNAK1, is important for efficient calcium signaling and NFAT activation in T cells through its ability to properly localize calcium channels at the plasma membrane [14]. In non-immune cells, AHNAK1 has also been shown to bind calcium channels with phospholipase Cγ (PLC-γ) and PKC.[1] Calcium binding proteins often quench much of the entering calcium, so linking these calcium effectors may be especially important when signals are induced by a weak calcium influx. |
| HOMER | Calcium signaling | Inhibition of NFAT activation | Another example of a scaffold protein that modulates calcium signaling is proteins of the HOMER family. The HOMER proteins have been shown to compete with calcineurin to bind to the N terminus of NFAT in activated T cells.[13] Through this competition, the HOMER proteins are able to reduce NFAT activation, which also reduces the production of the IL-2 cytokine.[13] In contrast, HOMER proteins have also been shown to positively regulate calcium signaling in neurons by linking the glutamate receptor with triphosphate receptors in the endoplasmic reticulum.[14] |
| Pellino | Innate Immune Signaling | Assembly of the TLR signalosome | Evidence exists that Pellino proteins function as scaffold proteins in the important innate immune signaling pathway, the Toll-like receptor (TLR) pathway. Much Pellino function is speculation; however, Pellino proteins can associate with IRAK1, TRAF6, and TAK1 following IL-1R activation, indicating that they may assemble and localize components of the TLR pathway near its receptor.[15][16] |
| NLRP | Innate Immune Signaling | Assembly of the inflammasome | The NLR family is a highly conserved and large family of receptors involved in innate immunity. The NLRP (NLR family, pyrine domain-containing) family of receptors function as scaffolds by assembling the inflammasome, a complex that leads to the secretion of pro-inflammatory cytokines such as IL-18 and IL-1β.[17] |
| DLG1 | T-cell receptor signaling | Assembly and localization of TCR signaling molecules, activation of p38 | DLG1 is highly conserved in immune cells and is important for T-cell activation in the periphery. It is recruited to the immunological synapse and links the ζ-chain of the T-cell receptor (TCR) to CBL, WASP, p38, LCK, VAV1, and ZAP70.[18][19][20][21] This data suggests that DLG1 plays a role in linking TCR signaling machinery with cytoskeleton regulators and also suggests a role in alternatively activating the p38 pathway. However, it is unclear to whether DLG1 positively or negatively regulates T-cell activation. |
| Spinophilin | Dendritic cell signaling | Assembly of DC immunological-synapse proteins | Spinophilin is involved in dendritic cell function specifically in the formation of immunological synapses. Spinophilin is recruited to the synapse following dendritic cell contact with a T cell. This recruitment seems to be important because without spinophilin, dendritic cells cannot activate T cells in vitro or in vivo.[22] How spinophilin facilitates antigen presentation in this case is still unknown though it is possible that spinophilin regulates the duration of cell contact in the synapse or regulates the recycling of co-stimulatory molecules in the cell like MHC molecules.[1] |
| Plant FLU regulatory protein[23] | Coordination of negative feedback during protochlorophyllide biosynthesis. | Assembly and localization of the pathway that turns of synthesis of highly toxic protochlorophyllide, a precursor of chlorophyll. | Synthesis of protochlorophyllide must be strictly regulated as its conversion into chlorophyll requires light. FLU regulatory protein is located in thylakoid membrane and only contains several protein-protein interaction sites without catalytic activity. Mutants lacking this protein overaccumulate protochlorophyllide in the darkness. The interaction partners are unknown. The protein underwent simplification during evolution. |
References
- 1 2 3 4 Shaw, A.S. and E.L. Filbert, Scaffold proteins and immune-cell signalling. Nat Rev Immunol, 2009. 9(1): p. 47–56.
- ↑ Choi, Kang-Yell; Satterberg, Brett; Lyons, David M.; Elion, Elaine A. (1994). "Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae". Cell. 78 (3): 499–512. doi:10.1016/0092-8674(94)90427-8. ISSN 0092-8674.
- 1 2 3 Levchenko, A., J. Bruck, and P.W. Sternberg, Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc Natl Acad Sci U S A, 2000. 97(11): p. 5818–23.
- ↑ Ferrell, J.E., Jr., What do scaffold proteins really do? Sci STKE, 2000. 2000(52): p. PE1.
- ↑ Burack, W.R. and A.S. Shaw, Signal transduction: hanging on a scaffold. Curr Opin Cell Biol, 2000. 12(2): p. 211–6.
- 1 2 Good, M., et al., The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell, 2009. 136(6): p. 1085–97.
- ↑ Wong, W. and J.D. Scott, AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol, 2004. 5(12): p. 959–70.
- 1 2 3 Locasale, J.W., A.S. Shaw, and A.K. Chakraborty, Scaffold proteins confer diverse regulatory properties to protein kinase cascades. Proc Natl Acad Sci U S A, 2007. 104(33): p. 13307–12.
- ↑ Uhlik, M.T., et al., Wiring diagrams of MAPK regulation by MEKK1, 2, and 3. Biochem Cell Biol, 2004. 82(6): p. 658–63.
- ↑ Greenwald, EC; Redden, JM; Dodge-Kafka, KL; Saucerman, JJ (24 January 2014). "Scaffold state switching amplifies, accelerates, and insulates protein kinase C signaling.". The Journal of Biological Chemistry. 289 (4): 2353–60. doi:10.1074/jbc.M113.497941. PMID 24302730.
- ↑ Claperon, A. and M. Therrien, KSR and CNK: two scaffolds regulating RAS-mediated RAF activation. Oncogene, 2007. 26(22): p. 3143–58.
- ↑ Muller, J., et al., C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol Cell, 2001. 8(5): p. 983–93.
- 1 2 Huang, G.N., et al., NFAT binding and regulation of T cell activation by the cytoplasmic scaffolding Homer proteins. Science, 2008. 319(5862): p. 476–81.
- ↑ Xiao, B., J.C. Tu, and P.F. Worley, Homer: a link between neural activity and glutamate receptor function. Curr Opin Neurobiol, 2000. 10(3): p. 370–4.
- ↑ Jiang, Z., et al., Pellino 1 is required for interleukin-1 (IL-1)-mediated signaling through its interaction with the IL-1 receptor-associated kinase 4 (IRAK4)-IRAK-tumor necrosis factor receptor-associated factor 6 (TRAF6) complex. J Biol Chem, 2003. 278(13): p. 10952–6.
- ↑ Yu, K.Y., et al., Cutting edge: mouse pellino-2 modulates IL-1 and lipopolysaccharide signaling. J Immunol, 2002. 169(8): p. 4075–8.
- ↑ Petrilli, V., et al., The inflammasome: a danger sensing complex triggering innate immunity. Curr Opin Immunol, 2007. 19(6): p. 615–22.
- ↑ Xavier, R., et al., Discs large (Dlg1) complexes in lymphocyte activation. J Cell Biol, 2004. 166(2): p. 173–8.
- ↑ Hanada, T., et al., Human homologue of the Drosophila discs large tumor suppressor binds to p56lck tyrosine kinase and Shaker type Kv1.3 potassium channel in T lymphocytes. J Biol Chem, 1997. 272(43): p. 26899–904.
- ↑ Round, J.L., et al., Scaffold protein Dlgh1 coordinates alternative p38 kinase activation, directing T cell receptor signals toward NFAT but not NF-kappaB transcription factors. Nat Immunol, 2007. 8(2): p. 154–61.
- ↑ Round, J.L., et al., Dlgh1 coordinates actin polymerization, synaptic T cell receptor and lipid raft aggregation, and effector function in T cells. J Exp Med, 2005. 201(3): p. 419–30.
- ↑ Bloom, O., et al., Spinophilin participates in information transfer at immunological synapses. J Cell Biol, 2008. 181(2): p. 203–11.
- ↑ Meskauskiene, R; Nater, M; Goslings, D; Kessler, F; op den Camp, R; Apel, K (2001). "FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana" (PDF). Proceedings of the National Academy of Sciences of the United States of America. 98 (22): 12826–31. doi:10.1073/pnas.221252798. PMC 60138
 . PMID 11606728.
. PMID 11606728.