SIGLEC
Siglecs (Sialic acid-binding immunoglobulin-type lectins) are cell surface proteins that bind sialic acid. They are found primarily on the surface of immune cells and are a subset of the I-type lectins. There are 14 different mammalian Siglecs, providing an array of different functions based on cell surface receptor-ligand interactions.
History
The first described candidate Siglec was Sialoadhesin (Siglec-1/CD169) a lectin-like adhesion protein on macrophages.[1] Parallel studies by Ajit Varki and colleagues on the previously cloned CD22 (a B cell surface protein involved in adhesion and activation) showed direct evidence for sialic acid recognition. The subsequent cloning of Sialoadhesin by Crocker revealed homology to CD22 (Siglec-2), CD33 (Siglec-3) and myelin-associated glycoprotein (MAG/Siglec-4), leading to the proposal for a family of "Sialoadhesins". Varki then suggested the term Siglec as a better alternative and as a subset of I-type (Ig-type) lectins. This nomenclature was agreed upon and has been adopted by almost all investigators working on these molecules (by convention, Siglecs are always capitalised.) Several additional Siglecs (Siglecs 5–12) have been identified in humans that are highly similar in structure to CD33 and so are collectively referred to as "CD33-related Siglecs".[2] Further Siglecs have been identified including Siglec-14 and Siglec-15. Some Siglecs are absolutely conserved between mice and humans including Sialoadhesin, CD22, MAG and Siglec-15. Others such as Siglec-8 and Siglec-9 have homologues in mice and rats (Siglec-F and Siglec-E respectively in both). Humans have a higher number of Siglecs than mice and so the numbering system was based on the human proteins.[3]
Structure
Siglecs are Type I transmembrane proteins where the NH3+-terminus is in the extracellular space and the COO−-terminus is cytosolic.[4] Each Siglec contains an N-terminal V-type immunoglobulin domain (Ig domain) which acts as the binding receptor for sialic acid. These lectins are placed into the group of I-type lectins because the lectin domain is an immunoglobulin fold. All Siglecs are extended from the cell surface by C2-type Ig domains which have no binding activity. Siglecs differ in the number of these C2-type domains.[3] As these proteins contain Ig domains, they are members of the Immunoglobulin superfamily (IgSF).
Most Siglecs, such as CD22 and the CD33-related family, contain ITIMs (Immunoreceptor tyrosine-based inhibitory motifs) in their cytosolic region.[4] These act to down-regulate signalling pathways involving phosphorylation, such as those induced by ITAMs (Immunoreceptor tyrosine-based activation motifs).[5] Some, however, like Siglec-14, contain positive amino acid residues that help dock ITAM-containing adaptor proteins such as DAP12.[6]
Ligand Binding
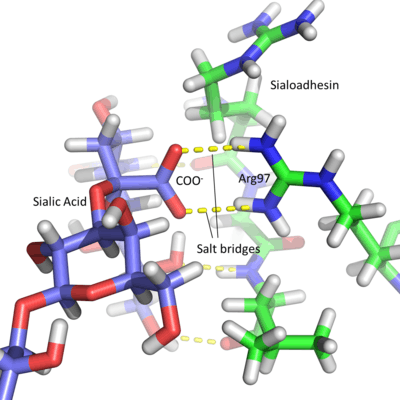
Due to the acidic nature of sialic acid, Siglec active sites contain a conserved arginine residue which is positively charged at physiological pH. This amino acid forms salt bridges with the carboxyl group of the sugar residue.[3] This is best seen in Sialoadhesin, where arginine at position 97 forms salt bridges with the COO− group of the sialic acid, producing a stable interaction.[7] Each lectin domain is specific for the linkage that connects sialic acid to the glycan. Sialic acid contains numerous hydroxyl groups which can be involved in the formation of glycosidic linkages. Most sialic acids are bonded via the 2, 3, 6 and occasionally 8 hydroxyl groups (number dependent on the carbon to which they are attached), in an α anomeric configuration. The specificity of each Siglec is due to different chemical interactions between the sugar ligand and the Siglec amino acids. The position in space of the individual groups on the sugar and the protein amino acids affects the sialic acid linkage to which each Siglec binds. For example, Sialoadhesin preferentially binds α2,3 linkages over α2,6 linkages.[7]
Function
The primary function of Siglecs is to bind glycans containing sialic acids. These receptor-glycan interactions can be used in cell adhesion, cell signalling and others. The function of Siglecs is limited to their cellular distribution. For example, MAG is found only on oligodendrocytes and schwann cells whereas Sialoadhesin is localised to macrophages.
Most Siglecs are short and do not extend far from the cell surface. This prevents most Siglecs from binding to other cells as mammalian cells are covered in sialic acid-containing glycans. This means that the majority of Siglecs only bind ligands on the surface of the same cell, so called cis -ligands, as they are "swamped" by glycans on the same cell. One exception is Sialoadhesin which contains 16 C2-Ig domains, producing a long, extended protein allowing it to bind trans-ligands, i.e. ligands found on other cells. Others, such as MAG, have also been shown to bind trans-ligands.
Signalling
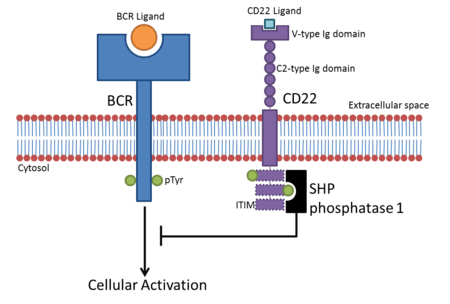
Due to their ITIM-containing cytoplasmic regions, most Siglecs interfere with cellular signalling, inhibiting immune cell activation. Once bound to their ligands, Siglecs recruit inhibitory proteins such as SHP phosphatases via their ITIM domains.[8] The tyrosine contained within the ITIM is phosphorylated after ligand binding and acts as a docking site for SH2 domain-containing proteins like SHP phosphatases. This leads to de-phosphorylation of cellular proteins, down-regulating activating signalling pathways.
Examples of negative signalling:
- CD22 is found on B cells. B cells become active when the B-cell receptor (BCR) binds to its cognate ligand. Once the BCR is bound to its ligand, the receptor auto-phosphorylates its cytoplasmic region (cytoplasmic tail). This leads to phosphorylation of the three ITIMs in CD22’s cytoplasmic tail, leading to the recruitment of SHP-1 which negatively regulates BCR-based cellular activation. This creates an activation threshold for B cell activation whereby transient activation of B cells is prevented.[9] CD22 inhibition of BCR signalling was originally thought to be sialic acid-binding-independent, but evidence suggests α2,6 sialic acid ligands are required for inhibition.[10]
- Siglec-7 is found on Natural Killer cells (NK cells). Siglec-7 leads to cellular inactivation once bound to its sialic acid-containing cognate ligand and is found in high levels on NK cell surfaces. It is used in cell-cell contacts, binding to sialylated glycans on target cells leading to inhibition of NK cell-dependent killing of the target cell. Mammalian cells contain high levels of sialic acid and so when NK cells bind so called "self-cells", they are not activated and do not kill host cells.
Siglec-14 contains an arginine residue in its transmembrane region.[11] This binds to the ITAM-containing DAP10 and DAP12 proteins. When bound to its ligand, Siglec-14 leads to activation of cellular signalling pathways via the DAP10 and DAP12 proteins.[4] These proteins up-regulate phosphorylation cascades involving numerous cellular proteins, leading to cellular activation. Siglec-14 appears to co-localise with Siglec-5, and as this protein inhibits cellular signalling pathways, co-ordinate opposing functions within immune cells.[11]
Phagocytosis and adhesion
Siglecs that can bind trans-ligands, such as Sialoadhesin, allow cell-cell interactions to take place. These glycan-Siglec interactions allow cells to bind one another, allowing signalling in some cases, or in the case of Sialoadhesin, pathogen uptake. Sialoadhesin's function was originally thought to be important in binding to red blood cells. Sialoadhesin lacks a cytosolic ITIM or a positive residue to bind ITAM-containing adaptors and so is thought not to influence signalling. Studies show that this protein is involved in phagocytosis of bacteria that contain highly sialylated glycan structures such as the lipopolysaccharide of Neisseria meningitidis.[12] Binding to these structures allows the macrophage to phagocytose these bacteria, clearing the system of pathogens.
Siglec-7 is also used in binding to pathogens such as Campylobacter jejuni. This occurs in a sialic acid-dependent manner and brings NK cells and monocytes, on which Siglec-7 is expressed, into contact with these bacteria.[13] The NK cell is then able to kill these foreign pathogens.
Knock-out studies
Knock-out studies are often used to uncover the function proteins have within a cell. Mice are often used as they express orthologous proteins of ours, or extremely similar homologues.
Some examples of knock-out Siglecs include:
- CD22: Walker & Smith conducted experiments with CD22 knock-outs and deletion mutants to discern CD22's function.[14] These mutant B cells did not infer any autoimmune disease, but they did see an increased production of autoantibodies due to the lack of BCR signalling inhibition, usually conducted by CD22. Autoantibodies are specific for self proteins and can lead to harm in the host. CD22 is normally up-regulated by lipopolysaccharide binding to Toll-like receptors. The mutant B cells can not up-regulate the mutant protein and so become hyper-sensitive in the presence of lipopolysaccharide. This means that the B cells overproduce antibodies when antibodies would not normally have been produced.
- MAG (Myelin-associated glycoprotein) is expressed on cells that form myelin sheaths (schwann cells and oligodendrocytes) around neurons. MAG binds to sialylated ligands on the neuron. Knock-out of MAG in the peripheral nervous system leads to decreased myelination of neurons. Knock-out of MAG in the central nervous system of mice does not appear to affect myelination, but the interaction between the myelin and the neuron does deteriorate with age. This leads to neurological defects as the action potential can not pass so rapidly down the length of the axon during nerual stimulation. Removing the ligand for MAG, by knocking-out the GalNAc transferase gene required for ligand formation, has similar effects to that of the MAG knock-out mice[15]
Human/Primate Siglecs
| Name | Cellular distribution[4] | Sialic acid linkage specificity[3] | No. of C2-Ig domains[4] | ITIM or positive residue[4] |
|---|---|---|---|---|
| Siglec-1 (Sialoadhesin) | Macrophages | α2,3>α2,6 | 16 | None |
| Siglec-2 (CD22) | B cells | α2,6 | 6 | ITIM |
| Siglec-3 (CD33) | Myeloid progenitors, Monocytes | α2,6>α2,3[16] | 1 | ITIM |
| Siglec-4 (MAG) | Myelin | α2,3>α2,6 | 4 | None |
| Siglec-5 | Neutrophils, Monocytes | α2,3 | 3 | ITIM |
| Siglec-6 | Trophoblasts | α2,6 | 2 | ITIM |
| Siglec-7 | NK cells | α2,8>α2,6>α2,3 | 2 | ITIM |
| Siglec-8 | Eosinophils | α2,3>α2,6 | 2 | ITIM |
| Siglec-9 | Monocytes, Neutrophils, Dendritic cells | α2,3=α2,6 (prefers sulfated residues) | 2 | ITIM |
| Siglec-10 | B cells | α2,3=α2,6 | 4 | ITIM |
| Siglec-11 | B cells | α2,8 | 4 | ITIM |
| Siglec-12[17] | Macrophages | No binding[3] | 2 | ITIM |
| Siglec-13[18] | Chimpanzee monocytes | |||
| Siglec-14 | Unknown | α2,6[11] | 2 | Arginine[11] |
| Siglec-15 | Unknown | α2,6[19] | 1 | Lysine[19] |
| Siglec-16[20] | Tissue macrophages | |||
| Siglec-17 [18] | NK cells | |||
This table briefly summarises the cellular distribution of each human/primate Siglec; the linkage specificity each has for sialic acid binding; the number of C2-Ig domains it contains; and whether it contains an ITIM or a positive residue to bind ITAM-containing adaptor proteins. References in the column headings correspond to all information displayed in that column, unless other references are shown. Siglec-12 information is referenced by[17] only, excluding the linkage specificity.
Reference List
- ↑ Crocker P; Gordon S (1986). "Properties and distribution of a lectin-like hemagglutinin differentially expressed by murine stromal tissue macrophages". J Exp Med. 164 (6): 1862–75. doi:10.1084/jem.164.6.1862. PMC 2188478
 . PMID 3783087.
. PMID 3783087. - ↑ Crocker P; Varki A (2001). "Siglecs in the immune system". Immunology. 103 (2): 137–45. doi:10.1046/j.0019-2805.2001.01241.x. PMC 1783234
 . PMID 11412300.
. PMID 11412300. - 1 2 3 4 5 Varki A; Angata T (2006). "Siglecs-the major subfamily of I-type lectins". Glycobiology. 16: 1R–27R. doi:10.1093/glycob/cwj008. PMID 16014749.
- 1 2 3 4 5 6 Crocker P; Paulson J; Varki A (2007). "Siglecs and their roles in the immune system". Nature Reviews Immunology. 7: 255–266. doi:10.1038/nri2056. PMID 17380156.
- ↑ Daëron M; Jaeger S; Du Pasquier L; Vivier E (2008). "Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future". Immunological Reviews. 224: 11–43. doi:10.1111/j.1600-065X.2008.00666.x. PMID 18759918.
- ↑ Pillai S; Netravali I; Cariappa A; Mattoo H (2012). "Siglecs and Immune Regulation". Annual Review of Immunology. 30: 357–392. doi:10.1146/annurev-immunol-020711-075018. PMID 22224769.
- 1 2 Hartnell A; Steel J; Turley H; Jones M; Jackson D; Crocker P (2001). "Characterization of human sialoadhesin, a sialic acid binding receptor expressed by resident and inflammatory macrophage populations". Blood. 97: 288–296. doi:10.1182/blood.V97.1.288. PMID 11133773.
- ↑ Avril T; Floyd H; Lopez F; Vivier E; Crocker P (2004). "The Membrane-Proximal Immunoreceptor Tyrosine-Based Inhibitory Motif is critical for the Inhibitory Signaling Mediated by Siglecs-7 and -9, CD33-Related Siglecs Expressed on Human Monocytes and NK Cells". The Journal of Immunology. 173: 6841–6849. doi:10.4049/jimmunol.173.11.6841. PMID 15557178.
- ↑ Nitschke L; Carsetti R; Ocker B; Köhler G; Lamers M (1997). "CD22 is a negative regulator of B-cell receptor signalling". Current Biology. 7: 133–143. doi:10.1016/S0960-9822(06)00057-1. PMID 9016707.
- ↑ Nitschke L; Tsubata T (2004). "Molecular interactions regulated BCR signal inhibition by CD22 and CD72". Trends in Immunology. 25: 543–550. doi:10.1016/j.it.2004.08.002. PMID 15364057.
- 1 2 3 4 Angata T; Hayakawa T; Wamanaka M; Warki A; Nahamura M (2006). "Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates". FASEB J. 20: 1964–1976. doi:10.1096/fj.06-5800com. PMID 17012248.
- ↑ Jones C; Virji M; Crocker P (2003). "Recognition of sialylated meningococcal lipopolysaccharide by Siglecs expressed on myeloid cells leads to enhanced bacterial uptake". Mol. Microbiol. 49: 1213–1225. doi:10.1046/j.1365-2958.2003.03634.x. PMID 12940982.
- ↑ Avril T; Wagner E; Willison H; Crocker P (2006). "Sialic Acid-Binding Immunoglobulin-Like Lectin 7 Mediates Selective Recognition of Sialylated Glycans Expressed on Campylobacter jejuni Lipooligosaccharides". Infection and Immunity. 74: 4133–4141. doi:10.1128/IAI.02094-05. PMID 16790787.
- ↑ Walker J; Smith K (2008). "CD22: an inhibitory enigma". Immunology. 123: 314–325. doi:10.1111/j.1365-2567.2007.02752.x. PMID 18067554.
- ↑ Taylor, Maureen E.; Drickamer, Kurt (2011). "Chapter 12: Glycobiology and Development". Introduction to Glycobiology (3rd ed.). Oxford University Press. pp. 228–235. ISBN 978-0-19-956911-3..
- ↑ Razi N; Varki A (1999). "Cryptic sialic acid binding lectins on human blood leukocytes can be unmasked by sialidase treatment or cellular activation". Glycobiology. 9: 1225–1234. doi:10.1093/glycob/9.11.1225. PMID 10536038.
- 1 2 Mitra N; Banda K; Altheide T; Schaffer L; Johnson-Pais T; Beuten J; Leach R; Angata T; Varki N; Varki A (2011). "SIGLEC12, a Human-specific Segragating (Pseudo)gene, Encodes a Signaling Molecule Expressed in Prostate Carcinomas". J. Biol. Chem. 286: 23003–23011. doi:10.1074/jbc.M111.244152. PMID 21555517.
- 1 2 Wang X; Mitra N; Secundino I; Banda K; Cruz P; Padler-Karavani V; Verhagen A; Reid C; Lari M; Rizzi E; Balsamo C; Corti G; De Bellis G; Longo L; NISC Comparative Sequencing Program; Beggs W; Caramelli D; Tishkoff SA; Hayakawa T; Green ED; Mullikin JC; Nizet V; Bui J; Varki A (Jun 19, 2012). "Specific inactivation of two immunomodulatory SIGLEC genes during human evolution". Proc Natl Acad Sci U S A. 109 (25): 9935–40. doi:10.1073/pnas.1119459109.
- 1 2 Angata T; Tabuchi Y; Nakamura K; Nakamura M (2007). "Siglec-15: an immune system Siglec conserved throughout vertebrate evolution". Glycobiology. 17: 838–846. doi:10.1093/glycob/cwm049. PMID 17483134.
- ↑ Cao H1, Lakner U, de Bono B, Traherne JA, Trowsdale J, Barrow AD. (Aug 2008). "SIGLEC16 encodes a DAP12-associated receptor expressed in macrophages that evolved from its inhibitory counterpart SIGLEC11 and has functional and non-functional alleles in humans". Eur J Immunol. 38 (8): 2303–15. doi:10.1002/eji.200738078.
External links
- Functional Glycomics Gateway, a collaboration between the Consortium for Functional Glycomics and Nature Publishing Group
- I-type Lectins (Siglecs) Research conducted at Imperial College London providing another overview of I-type Lectins