Restriction fragment length polymorphism
In molecular biology, restriction fragment length polymorphism, or RFLP, is a technique that exploits variations in homologous DNA sequences. It refers to a difference between samples of homologous DNA molecules from differing locations of restriction enzyme sites, and to a related laboratory technique by which these segments can be illustrated. In RFLP analysis, the DNA sample is broken into pieces and (digested) by restriction enzymes and the resulting restriction fragments are separated according to their lengths by gel electrophoresis. Although now largely obsolete due to the rise of inexpensive DNA sequencing technologies, RFLP analysis was the first DNA profiling technique inexpensive enough to see widespread application. RFLP analysis was an important tool in genome mapping, localization of genes for genetic disorders, determination of risk for disease, and paternity testing.
Analysis (technology)
The basic technique for the detecting of RFLPs involves fragmenting a sample of DNA by a restriction enzyme, which can recognize and cut DNA wherever a specific short sequence occurs, in a process known as a restriction digest. The resulting DNA fragments are then separated by length through a process known as agarose gel electrophoresis, and transferred to a membrane via the Southern blot procedure. Hybridization of the membrane to a labeled DNA probe then determines the length of the fragments which are complementary to the probe. An RFLP occurs when the length of a detected fragment varies between individuals. Each fragment length is considered an allele, and can be used in genetic analysis.
RFLP analysis may be subdivided into single- (SLP) and multi-locus probe (MLP) paradigms. Usually, the SLP method is preferred over MLP because it is more sensitive, easier to interpret and capable of analyzing mixed-DNA samples. Moreover, data can be generated even when the DNA is degraded (e.g. when it is found in bone remains.)
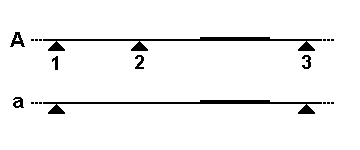


Examples
There are two common mechanisms by which the size of a particular restriction fragment can vary. In the first schematic, a small segment of the genome is being detected by a DNA probe (thicker line). In allele "A", the genome is cleaved by a restriction enzyme at three nearby sites (triangles), but only the rightmost fragment will be detected by the probe. In allele "a", restriction site 2 has been lost by a mutation, so the probe now detects the larger fused fragment running from sites 1 to 3. The second diagram shows how this fragment size variation would look on a Southern blot, and how each allele (two per individual) might be inherited in members of a family.
In the third schematic, the probe and restriction enzyme are chosen to detect a region of the genome that includes a variable number tandem repeat segment (boxes in schematic diagram). In allele "c" there are five repeats in the VNTR, and the probe detects a longer fragment between the two restriction sites. In allele "d" there are only two repeats in the VNTR, so the probe detects a shorter fragment between the same two restriction sites. Other genetic processes, such as insertions, deletions, translocations, and inversions, can also lead to RFLPs. RFLP tests require much bigger samples of DNA than do short tandem repeat (STR) tests.
Applications
Analysis of RFLP variation in genomes was a vital tool in genome mapping and genetic disease analysis. If researchers were trying to initially determine the chromosomal location of a particular disease gene, they would analyze the DNA of members of a family afflicted by the disease, and look for RFLP alleles that show a similar pattern of inheritance as that of the disease (see Genetic linkage). Once a disease gene was localized, RFLP analysis of other families could reveal who was at risk for the disease, or who was likely to be a carrier of the mutant genes.
RFLP analysis was also the basis for early methods of genetic fingerprinting, useful in the identification of samples retrieved from crime scenes, in the determination of paternity, and in the characterization of genetic diversity or breeding patterns in animal populations.
Alternatives
The technique for RFLP analysis is, however, slow and cumbersome. It requires a large amount of sample DNA, and the combined process of probe labeling, DNA fragmentation, electrophoresis, blotting, hybridization, washing, and autoradiography could take up to a month to complete. A limited version of the RFLP method that used oligonucleotide probes was reported in 1985.[1] Fortunately, the results of the Human Genome Project have largely replaced the need for RFLP mapping, and the identification of many single-nucleotide polymorphisms (SNPs) in that project (as well as the direct identification of many disease genes and mutations) has replaced the need for RFLP disease linkage analysis (see SNP genotyping). The analysis of VNTR alleles continues, but is now usually performed by polymerase chain reaction (PCR) methods. For example, the standard protocols for DNA fingerprinting involve PCR analysis of panels of more than a dozen VNTRs.
RFLP is still a technique used in marker assisted selection. Terminal restriction fragment length polymorphism (TRFLP or sometimes T-RFLP) is a molecular biology technique initially developed for characterizing bacterial communities in mixed-species samples. The technique has also been applied to other groups including soil fungi.
TRFLP works by PCR amplification of DNA using primer pairs that have been labeled with fluorescent tags. The PCR products are then digested using RFLP enzymes and the resulting patterns visualized using a DNA sequencer. The results are analyzed either by simply counting and comparing bands or peaks in the TRFLP profile, or by matching bands from one or more TRFLP runs to a database of known species. The technique is similar in some aspects to DGGE or TGGE.
The sequence changes directly involved with an RFLP can also be analyzed more quickly by PCR. Amplification can be directed across the altered restriction site, and the products digested with the restriction enzyme. This method has been called Cleaved Amplified Polymorphic Sequence (CAPS). Alternatively, the amplified segment can be analyzed by Allele specific oligonucleotide (ASO) probes, a process that can often be done by a simple Dot blot.
See also
References
- ↑ Saiki, RK; Scharf S; Faloona F; Mullis KB; Erlich HA; Arnheim N (Dec 20, 1985). "Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia". Science. 230 (4732): 1350–1354. doi:10.1126/science.2999980. PMID 2999980.
External links
- http://www.bio.davidson.edu/courses/genomics/method/RFLP.html
- http://www.ncbi.nlm.nih.gov/projects/genome/probe/doc/TechRFLP.shtml
- http://highered.mcgraw-hill.com/olc/dl/120078/bio20.swf