Quorum sensing
Quorum sensing is a system of stimuli and response correlated to population density. Many species of bacteria use quorum sensing to coordinate gene expression according to the density of their local population. In similar fashion, some social insects use quorum sensing to determine where to nest. In addition to its function in biological systems, quorum sensing has several useful applications for computing and robotics.
Quorum sensing can function as a decision-making process in any decentralized system, as long as individual components have: (a) a means of assessing the number of other components they interact with and (b) a standard response once a threshold number of components is detected.
Bacteria
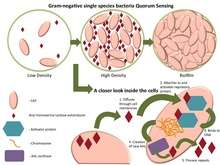
Some of the best-known examples of quorum sensing come from studies of bacteria. Bacteria use quorum sensing to coordinate certain behaviors such as biofilm formation, virulence, and antibiotic resistance, based on the local density of the bacterial population. Quorum sensing can occur within a single bacterial species as well as between diverse species, and can regulate a host of different processes, in essence, serving as a simple indicator of population density or the diffusion rate of the cell's immediate environment. A variety of different molecules can be used as signals. Common classes of signaling molecules are oligopeptides in Gram-positive bacteria, N-acyl homoserine lactones (AHL) in Gram-negative bacteria, and a family of autoinducers known as autoinducer-2 (AI-2) in both Gram-negative and Gram-positive bacteria.[1]
Mechanism
Bacteria that use quorum sensing constitutively produce and secrete certain signaling molecules (called autoinducers or pheromones). These bacteria also have a receptor that can specifically detect the signaling molecule (inducer). When the inducer binds the receptor, it activates transcription of certain genes, including those for inducer synthesis. There is a low likelihood of a bacterium detecting its own secreted inducer. Thus, in order for gene transcription to be activated, the cell must encounter signaling molecules secreted by other cells in its environment. When only a few other bacteria of the same kind are in the vicinity, diffusion reduces the concentration of the inducer in the surrounding medium to almost zero, so the bacteria produce little inducer. However, as the population grows, the concentration of the inducer passes a threshold, causing more inducer to be synthesized. This forms a positive feedback loop, and the receptor becomes fully activated. Activation of the receptor induces the up-regulation of other specific genes, causing all of the cells to begin transcription at approximately the same time. This coordinated behavior of bacterial cells can be useful in a variety of situations. For instance, the bioluminescent luciferase produced by Vibrio fischeri would not be visible if it were produced by a single cell. By using quorum sensing to limit the production of luciferase to situations when cell populations are large, V. fischeri cells are able to avoid wasting energy on the production of useless product.
In recent research, the direction for activating or deactivating nature of a wave of gene expression is predicted experimentally which control bacterial populations subject to a diffusing autoinducer signal. On the other hand, it has been observed that the accumulation of the quorum sensing molecules leads to a negative diffusion coefficient in the solution of governing differential equation.[2]
Examples
Aliivibrio fischeri
Quorum sensing was first observed in Aliivibrio fischeri, a bioluminescent bacterium that lives as a mutualistic symbiont in the photophore (or light-producing organ) of the Hawaiian bobtail squid.[3] When A. fischeri cells are free-living (or planktonic), the autoinducer is at low concentration, and, thus, cells do not luminesce. However, when they are highly concentrated in the photophore (about 1011 cells/ml), transcription of luciferase is induced, leading to bioluminescence.
Escherichia coli
In the Gram-negative bacterium Escherichia coli (E. coli), cell division may be partially regulated by AI-2-mediated quorum sensing. This species uses AI-2, which is produced and processed by the lsr operon. Part of it encodes an ABC transporter, which imports AI-2 into the cells during the early stationary (latent) phase of growth. AI-2 is then phosphorylated by the LsrK kinase, and the newly produced phospho-AI-2 can be either internalized or used to suppress LsrR, a repressor of the lsr operon (thereby activating the operon). Transcription of the lsr operon is also thought to be inhibited by dihydroxyacetone phosphate (DHAP) through its competitive binding to LsrR. Glyceraldehyde 3-phosphate has also been shown to inhibit the lsr operon through cAMP-CAPK-mediated inhibition. This explains why, when grown with glucose, E. coli will lose the ability to internalize AI-2 (because of catabolite repression). When grown normally, AI-2 presence is transient.
E. coli and Salmonella enterica do not produce AHL signals commonly found in other Gram-negative bacteria. However, they have a receptor that detects AHLs from other bacteria and change their gene expression in accordance with the presence of other "quorate" populations of Gram-negative bacteria.[4]
Salmonella enterica
Salmonella encodes a LuxR homolog, SdiA, but does not encode an AHL synthase. SdiA detects AHLs produced by other species of bacteria including Aeromonas hydrophila, Hafnia alvei, and Yersinia enterocolitica.[5] When AHL is detected, SdiA regulates the rck operon on the Salmonella virulence plasmid (pefI-srgD-srgA-srgB-rck-srgC) and a single gene horizontal acquisition in the chromosome srgE.[6][7] Salmonella does not detect AHL when passing through the gastrointestinal tracts of several animal species, suggesting that the normal microbiota does not produce AHLs. However, SdiA does become activated when Salmonella transits through turtles colonized with Aeromonas hydrophila or mice infected with Yersinia enterocolitica.[8][9] Therefore, Salmonella appears to use SdiA to detect the AHL production of other pathogens rather than the normal gut flora.
Pseudomonas aeruginosa
The opportunistic pathogen Pseudomonas aeruginosa uses quorum sensing to coordinate the formation of biofilms, swarming motility, exopolysaccharide production, virulence, and cell aggregation.[10] These bacteria can grow within a host without harming it, until they reach a threshold concentration. Then they become aggressive, developing to the point at which their numbers are sufficient to overcome the host's immune system, and form a biofilm, leading to disease within the host as the biofilm is a protective layer encasing the bacteria population. Another form of gene regulation that allows the bacteria to rapidly adapt to surrounding changes is through environmental signaling. Recent studies have discovered that anaerobiosis can significantly impact the major regulatory circuit of quorum sensing. This important link between quorum sensing and anaerobiosis has a significant impact on production of virulence factors of this organism.[11] Garlic and ginseng experimentally block quorum sensing in Pseudomonas aeruginosa.[12] It is hoped that the therapeutic enzymatic degradation of the signaling molecules will prevent the formation of such biofilms and possibly weaken established biofilms. Disrupting the signalling process in this way is called quorum sensing inhibition.
Acinetobacter sp.
It has recently been found that Acinetobacter sp. also show quorum sensing activity. This bacterium, an emerging pathogen, produces AHLs.[13] Interestingly, Acinetobacter sp. shows both quorum sensing and quorum quenching activity. It produces AHLs and also, it can degrade the AHL molecules as well.[13]
Aeromonas sp.
This bacterium was previously considered a fish pathogen, but it has recently emerged as a human pathogen.[14] Aeromonas sp. have been isolated from various infected sites from patients (bile, blood, peritoneal fluid, pus, stool and urine). All isolates produced the two principal AHLs, N-butanoylhomoserine lactone (C4-HSL) and N-hexanoyl homoserine lactone (C6-HSL). It has been documented that Aeromonas sobria has produced C6-HSL and two additional AHLs with N-acyl side chain longer than C6.[15]
Yersinia
The YenR and YenI proteins produced by the gammaproteobacterium Yersinia enterocolitica are similar to Aliivibrio fischeri LuxR and LuxI.[16][17] YenR activates the expression of a small non-coding RNA, YenS. YenS inhibits YenI expression and acylhomoserine lactone production.[18] YenR/YenI/YenS are involved in the control of swimming and swarming motility.[17][18]
Molecules involved in quorum sensing
Three-dimensional structures of proteins involved in quorum sensing were first published in 2001, when the crystal structures of three LuxS orthologs were determined by X-ray crystallography.[19] In 2002, the crystal structure of the receptor LuxP of Vibrio harveyi with its inducer AI-2 (which is one of the few biomolecules containing boron) bound to it was also determined.[20] Many bacterial species, including E. coli, an enteric bacterium and model organism for Gram-negative bacteria, produce AI-2. A comparative genomic and phylogenetic analysis of 138 genomes of bacteria, archaea, and eukaryotes found that "the LuxS enzyme required for AI-2 synthesis is widespread in bacteria, while the periplasmic binding protein LuxP is present only in Vibrio strains," leading to the conclusion that either "other organisms may use components different from the AI-2 signal transduction system of Vibrio strains to sense the signal of AI-2 or they do not have such a quorum sensing system at all."[21] Farnesol is used by the fungus Candida albicans as a quorum sensing molecule that inhibits filamentation.[22]
A database of quorum-sensing peptides is available under the name Quorumpeps.[23][24]
Certain bacteria can produce enzymes called lactonases that can target and inactivate AHLs. Researchers have developed novel molecules which block the signalling receptors of bacteria. mBTL is a compound that has been shown to inhibit quorum sensing and decrease the amount of cell death by a significant amount.[25] Additionally, researchers are also examining the role of natural compounds (such as caffeine) as potential quorum sensing inhibitors.[26] Research in this area has been promising and could lead to the development of natural compounds as effective therapeutics.
Evolution
Sequence analysis
The majority of quorum sensing systems that fall under the "two-gene" (an autoinducer synthase coupled with a receptor molecule) paradigm as defined by the Vibrio fischeri system occur in the Gram-negative Proteobacteria. A comparison between the Proteobacteria phylogeny as generated by 16S ribosomal RNA sequences and phylogenies of LuxI-, LuxR-, or LuxS-homologs shows a notably high level of global similarity. Overall, the quorum sensing genes seem to have diverged along with the Proteobacteria phylum as a whole. This indicates that these quorum sensing systems are quite ancient, and arose very early in the Proteobacteria lineage.[27][28]
Although examples of horizontal gene transfer are apparent in LuxI, LuxR, and LuxS phylogenies, they are relatively rare. This result is in line with the observation that quorum sensing genes tend to control the expression of a wide array of genes scattered throughout the bacterial chromosome. A recent acquisition by horizontal gene transfer would be unlikely to have integrated itself to this degree. Given that the majority of autoinducer–synthase/receptor occurs in tandem in bacterial genomes, it is also rare that they switch partners and so pairs tend to co-evolve.[28]
The phylogeny of quorum sensing genes in Gammaproteobacteria (which includes Pseudomonas aeruginosa and Escherichia coli) is especially interesting. The LuxI/LuxR genes form a functional pair, with LuxI as the auto-inducer synthase and LuxR as the receptor. Gamma Proteobacteria are unique in possessing quorum sensing genes, which, although functionally similar to the LuxI/LuxR genes, have a markedly divergent sequence.[28] This family of quorum-sensing homologs may have arisen in the gamma Proteobacteria ancestor, although the cause of their extreme sequence divergence yet maintenance of functional similarity has yet to be explained. In addition, species that employ multiple discrete quorum sensing systems are almost all members of the gamma Proteobacteria, and evidence of horizontal transfer of quorum sensing genes is most evident in this class.[27][28]
Controversy
As quorum sensing implies a cooperative behavior, this concept has been challenged by the evolutionary implication of cooperative cheaters. This is circumvented by the concept of diffusion sensing, which has been an alternative and complementary model to quorum sensing.[29] However, both explanations face the problems of signalling in either complex (multiple species sharing the same space) or simple (one single cell confined to a limited volume) environments where the spatial distribution of the cells can be more important for sensing than the cell population density. A new model, efficiency sensing, which takes into account both problematics, population density and spatial confinement, has been proposed as an alternative.[30] One of the probable reasons for controversy is that current terminologies (quorum sensing, diffusion sensing, efficiency sensing) arise from different models of how natural selection has acted to shape and maintain the process. Since each of the various models does apply under some circumstances but not others, a sensible resolution to these controversies could be to return the terminology of the process to autoinduction, as originally described by Hastings and coworkers, as this term does not imply understanding of the function of the process.
Interaction of quorum-sensing molecules with mammalian cells and its medical applications
Next to the potential antimicrobial functionality, quorum-sensing derived molecules, especially the peptides, are being investigated for their use in other therapeutic domains as well, including immunology, central nervous system disorders and oncology. Quorum-sensing peptides have been demonstrated to interact with cancer cells, as well as to permeate the blood-brain barrier permeation reaching the brain parenchyma[31][32][33][34]
Archaea
Examples
Methanosaeta harundinacea 6Ac
Methanosaeta harundinacea 6Ac, a methanogenic archaeon, produces carboxylated acyl homoserine lactone compounds that facilitate the transition from growth as short cells to growth as filaments.[35]
Quorum quenching
Quorum quenching is the process of preventing quorum sensing by disrupting the signaling.[36] This may be achieved by degrading the signalling molecule.[37][38] Using a KG medium, quorum quenching bacteria can be readily isolated from various environments including that which has previously been considered as unculturable.[38] Recently, a well-studied quorum quenching bacterium has been isolated and its AHL degradation kinetic has been studied by using rapid resolution liquid chromatography (RRLC).[39]
Social insects
Social insect colonies are an excellent example of a decentralized system, because no individual is in charge of directing or making decisions for the colony. Several groups of social insects have been shown to use quorum sensing in a process that resembles collective decision-making.
Examples
Ants
Colonies of the ant Temnothorax albipennis nest in small crevices between rocks. When the rocks shift and the nest is broken up, these ants must quickly choose a new nest to move into. During the first phase of the decision-making process, a small portion of the workers leave the destroyed nest and search for new crevices. When one of these scout ants finds a potential nest, she assesses the quality of the crevice based on a variety of factors including the size of the interior, the number of openings (based on light level), and the presence or absence of dead ants.[40][41] The worker then returns to the destroyed nest, where she waits for a short period before recruiting other workers to follow her to the nest that she has found, using a process called tandem running. The waiting period is inversely related to the quality of the site; for instance, a worker that has found a poor site will wait longer than a worker that encountered a good site.[42] As the new recruits visit the potential nest site and make their own assessment of its quality, the number of ants visiting the crevice increases. During this stage, ants may be visiting many different potential nests. However, because of the differences in the waiting period, the number of ants in the best nest will tend to increase at the greatest rate. Eventually, the ants in this nest will sense that the rate at which they encounter other ants has exceeded a particular threshold, indicating that the quorum number has been reached.[43] Once the ants sense a quorum, they return to the destroyed nest and begin rapidly carrying the brood, queen, and fellow workers to the new nest. Scouts that are still tandem-running to other potential sites are also recruited to the new nest, and the entire colony moves. Thus, although no single worker may have visited and compared all of the available options, quorum sensing enables the colony as a whole to quickly make good decisions about where to move.
Honey bees
Honey bees (Apis mellifera) also use quorum sensing to make decisions about new nest sites. Large colonies reproduce through a process called budding, in which the queen leaves the hive with a portion of the workers to form a new nest elsewhere. After leaving the nest, the workers form a swarm that hangs from a branch or overhanging structure. This swarm persists during the decision-making phase until a new nest site is chosen.
The quorum sensing process in honey bees is similar to the method used by Temnothorax ants in several ways. A small portion of the workers leave the swarm to search out new nest sites, and each worker assesses the quality of the cavity it finds. The worker then returns to the swarm and recruits other workers to her cavity using the honey bee waggle dance. However, instead of using a time delay, the number of dance repetitions the worker performs is dependent on the quality of the site. Workers that found poor nests stop dancing sooner, and can therefore be recruited to the better sites. Once the visitors to a new site sense that a quorum number (usually 10–20 bees) has been reached, they return to the swarm and begin using a new recruitment method called piping. This vibration signal causes the swarm to take off and fly to the new nest location. In an experimental test, this decision-making process enabled honey bee swarms to choose the best nest site in four out of five trials.[44][45]
Computing and robotics
Quorum sensing can be a useful tool for improving the function of self-organizing networks such as the SECOAS (Self-Organizing Collegiate Sensor) environmental monitoring system. In this system, individual nodes sense that there is a population of other nodes with similar data to report. The population then nominates just one node to report the data, resulting in power savings.[46] Ad-hoc wireless networks can also benefit from quorum sensing, by allowing the system to detect and respond to network conditions.[47]
Quorum sensing can also be used to coordinate the behavior of autonomous robot swarms. Using a process similar to that used by Temnothorax ants, robots can make rapid group decisions without the direction of a controller.[48]
See also
- Cell signaling
- Collective behavior
- Interspecies quorum sensing
- Microbial intelligence
- Swarm intelligence
References
- ↑ Miller, M.B.; Bassler, B.L. (2001). "Quorum sensing in bacteria". Annu. Rev. Microbiol. 55 (1): 165–99. doi:10.1146/annurev.micro.55.1.165. PMID 11544353.
- ↑ Sarangam Majumdar; Subhoshmita Mondal (2016). "Conversation game: talking bacteria". Journal of Cell Communication and Signaling: 1–5. doi:10.1007/s12079-016-0333-y. PMID 27278085.
- ↑ Nealson, K.; Platt, T.; Hastings, J.W. (1970). "The cellular control of the synthesis and activity of the bacterial luminescent system". Journal of Bacteriology. 104 (1): 313–22. PMC 248216
 . PMID 5473898.
. PMID 5473898. - ↑ Ahmer, B.M. (May 2004). "Cell-to-cell signalling in Escherichia coli and Salmonella enterica". Mol. Microbiol. 52 (4): 933–45. doi:10.1111/j.1365-2958.2004.04054.x. PMID 15130116.
- ↑ Michael, B.; Smith, J.N.; Swift, S.; Heffron, F.; Ahmer, B.M. (October 2001). "SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities". J. Bacteriol. 183 (19): 5733–42. doi:10.1128/JB.183.19.5733-5742.2001. PMC 95466
 . PMID 11544237.
. PMID 11544237. - ↑ Ahmer, B.M.; van Reeuwijk, J.; Timmers, C.D.; Valentine, P.J.; Heffron, F. (March 1998). "Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid". J. Bacteriol. 180 (5): 1185–93. PMC 107006
 . PMID 9495757.
. PMID 9495757. - ↑ Smith, J.N.; Ahmer, B.M. (February 2003). "Detection of other microbial species by Salmonella: expression of the SdiA regulon". J. Bacteriol. 185 (4): 1357–66. doi:10.1128/JB.185.4.1357-1366.2003. PMC 142872
 . PMID 12562806.
. PMID 12562806. - ↑ Smith, J.N.; Dyszel, J.L.; Soares, J.A.; et al. (2008). Ausubel, Frederick M., ed. "SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles". PLoS ONE. 3 (7): e2826. doi:10.1371/journal.pone.0002826. PMC 2475663
 . PMID 18665275.
. PMID 18665275. - ↑ Dyszel, J.L.; Smith, J.N.; Lucas, D.E.; et al. (January 2010). "Salmonella enterica serovar Typhimurium can detect acyl homoserine lactone production by Yersinia enterocolitica in mice". J. Bacteriol. 192 (1): 29–37. doi:10.1128/JB.01139-09. PMC 2798265
 . PMID 19820103.
. PMID 19820103. - ↑ Lewis Sauer, K.; Camper, A.; Ehrlich, G.; Costerton, J.; Davies, D. (2002). "Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm". Journal of Bacteriology. 184 (4): 1140–54. doi:10.1128/jb.184.4.1140-1154.2002. ISSN 0021-9193. PMC 134825
 . PMID 11807075.
. PMID 11807075. - ↑ Cornelis, P. (ed.) (2008). Pseudomonas: Genomics and Molecular Biology (1st ed.). Caister Academic Press. ISBN 1-904455-19-0.
- ↑ Thomas Bjarnsholt; Peter Østrup Jensen; Thomas B. Rasmussen; Lars Christophersen; Henrik Calum; Morten Hentzer; Hans-Petter Hougen; Jørgen Rygaard; Claus Moser; Leo Eberl; Niels Høiby & Michael Givskov (2005). "Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections". Microbiology. 151 (4): 3873–80. doi:10.1099/mic.0.27955-0. PMID 16339933.
- 1 2 Kok Gan, Chan; Atkinson, Steve; Mathee, Kalai; Sam, Choon-Kook; Chhabra, Siri Ram; Camara, Miguel; Koh, Chong-Lek & Williams, Paul (2011). "Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: Co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia". BMC Microbiology. 11 (1): 51. doi:10.1186/1471-2180-11-51. PMC 3062576
 . PMID 21385437.
. PMID 21385437. - ↑ "Emerging Aeromonas Species Infections and Their Significance in Public Health". ScientificWorldJournal. 2012: 625023. 2012. doi:10.1100/2012/625023. PMC 3373137
 . PMID 22701365.
. PMID 22701365. - ↑ Kok Gan, Chan; Puthucheary, Savithri D.; Chan, Xin-Yue; Yin, Wai-Fong; Wong, Cheng-Siang; Too, Wah-Seng See & Chua, Kek-Heng (2010). "Quorum sensing in Aeromonas species isolated from patients in Malaysia". Current Microbiology. 62 (1): 167–72. doi:10.1007/s00284-010-9689-z. PMID 20544198.
- ↑ Throup, JP; Camara, M; Briggs, GS; Winson, MK; Chhabra, SR; Bycroft, BW; Williams, P; Stewart, GS (July 1995). "Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules.". Molecular Microbiology. 17 (2): 345–56. doi:10.1111/j.1365-2958.1995.mmi_17020345.x. PMID 7494483.
- 1 2 Atkinson, S; Chang, CY; Sockett, RE; Cámara, M; Williams, P (February 2006). "Quorum sensing in Yersinia enterocolitica controls swimming and swarming motility.". Journal of Bacteriology. 188 (4): 1451–61. doi:10.1128/JB.188.4.1451-1461.2006. PMC 1367215
 . PMID 16452428.
. PMID 16452428. - 1 2 Tsai, CS; Winans, SC (April 2011). "The quorum-hindered transcription factor YenR of Yersinia enterocolitica inhibits pheromone production and promotes motility via a small non-coding RNA.". Molecular Microbiology. 80 (2): 556–71. doi:10.1111/j.1365-2958.2011.07595.x. PMID 21362062.
- ↑ Lewis, H.A.; Furlong, E.B.; Laubert, B.; Eroshkina, G.A.; Batiyenko, Y.; Adams, J.M.; Bergseid, M.G.; Marsh, C.D.; Peat, T.S.; Sanderson, W.E.; Sauder, J.M.; Buchanan, S.G. (2001). "A structural genomics approach to the study of quorum sensing: Crystal structures of three LuxS orthologs". Structure. 9 (6): 527–37. doi:10.1016/S0969-2126(01)00613-X. PMID 11435117.
- ↑ Chen, X.; Schauder, S.; Potier, N.; Van Dorsselaer, A.; Pelczer, I.; Bassler, B.; Hughson, F. (2002). "Structural identification of a bacterial quorum-sensing signal containing boron" (PDF). Nature. 415 (6871): 545–9. doi:10.1038/415545a. PMID 11823863.
- ↑ Sun, J.; Daniel, R.; Wagner-Döbler, I.; Zeng, A.P. (2004). "Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways". BMC Evol. Biol. 4 (1): 36. doi:10.1186/1471-2148-4-36. PMC 524169
 . PMID 15456522.
. PMID 15456522. - ↑ Jacob M. Hornby. "Quorum Sensing in the Dimorphic Fungus Candida albicans Is Mediated by Farnesol". asm.org. Retrieved 29 July 2015.
- ↑ "Quorumpeps database: chemical space, microbial origin and functionality of quorum sensing peptides". Nucleic Acids Research. 41: 1–5. 2012. doi:10.1093/nar/gks1137.
- ↑ "Exploring the chemical space of quorum sensing peptides". Biopolymers. 104 (5): 544–51. Sep 2015. doi:10.1002/bip.22649.
- ↑ O'Loughlin, CT.; et al. (2013). "A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation". PNAS. 110 (44): 17981–6. doi:10.1073/pnas.1316981110.
- ↑ Norizan, S.; Chan, K. (2013). "Caffeine as a potential quorum-sensing inhibitor". Sensors. 13 (4): 5117–5129. doi:10.3390/s130405117.
- 1 2 Gray, K.M.; Garey, J.R. (2001). "The evolution of bacterial LuxI and LuxR quorum sensing regulators". Microbiology. 147 (Pt 8): 2379–87. doi:10.1099/00221287-147-8-2379. PMID 11496014.
- 1 2 3 4 Lerat, E.; Moran, N.A. (2004). "Evolutionary history of quorum-sensing systems in bacteria". Molecular Biology and Evolution. 21 (5): 903–13. doi:10.1093/molbev/msh097. PMID 15014168.
- ↑ Redfield, RJ (2002). "Is quorum sensing a side effect of diffusion sensing". Trends in Microbiology. 10 (8): 365–370. doi:10.1016/s0966-842x(02)02400-9. PMID 12160634.
- ↑ Hense, B.A.; Kuttler, C.; Müller, J.; Rothballer, M.; Hartmann, A.; Kreft, J.U. (2007). "Does efficiency sensing unify diffusion and quorum sensing?". Nature Reviews Microbiology. 5 (3): 230–39. doi:10.1038/nrmicro1600. PMID 17304251.
- ↑ Wynendaele, E.; Pauwels, E.; Van de Wiele, C.; Burvenich, C.; De Spiegeleer, B. (2012). "The potential role of quorum-sensing peptides in oncology". Medical Hypotheses. 78: 814–817. doi:10.1016/j.mehy.2012.03.018.
- ↑ De Spiegeleer, B; Verbeke, F; D'Hondt, M; Hendrix, A; Van De Wiele, C; Burvenich, C; et al. (2015). "The Quorum Sensing Peptides PhrG, CSP and EDF Promote Angiogenesis and Invasion of Breast Cancer Cells In Vitro". PLoS ONE. 10 (3): e0119471. doi:10.1371/journal.pone.0119471.
- ↑ "Crosstalk between the microbiome and cancer cells by quorum sensing peptides". Peptides. 64: 40–8. Feb 2015. doi:10.1016/j.peptides.2014.12.009.
- ↑ Wynendaele, E; Verbeke, F; Stalmans, S; Gevaert, B; Janssens, Y; Van De Wiele, C; Peremans, K; Burvenich, C; De Spiegeleer, B (Nov 2015). "Quorum Sensing Peptides Selectively Penetrate the Blood-Brain Barrier". PLOS ONE. 10 (11): e0142071. doi:10.1371/journal.pone.0142071.
- ↑ Zhang, G. et al. (2012) Acyl homoserine lactone-based quorum sensing in a methanogenic archaeon. The ISME Journal. advanced online publication
- ↑ Dessaux, Y.; Chapelle, E.; Faure, D.; (2011). Quorum sensing and quorum quenching in soil ecosystems. In: G. Witzany (ed). Biocommunication in Soil Microorganisms. Springer; 339-367. ISBN 978-3-642-14511-7.
- ↑ Kok Gan, Chan; Atkinson, Steve; Kalai Mat hee; Choon-Kook Sam; Siri Ram Chhabra; Miguel Camara; Chong-Lek Koh & Paul Williams (2011). "Characterization of N-Acylhomoserine Lactone-Degrading Bacteria Associated with the Zingiber officinale (ginger) rhizosphere: Co-existence of Quorum Quenching and Quorum Sensing in Acinetobacter and Burkholderia". BMC Microbiology. 11 (1): 51. doi:10.1186/1471-2180-11-51. PMC 3062576
 . PMID 21385437.
. PMID 21385437. - 1 2 Kok Gan, Chan; Wai-Fong Yin; Choon-Kook Sam (2009). "A novel medium for the isolation of N-acylhomoserine lactone-degrading bacteria". Journal of Industrial Microbiology & Biotechnology. 36 (2): 247–51. doi:10.1007/s10295-008-0491-x.
- ↑ Kok Gan, Chan; Wong, Cheng-Siang; Yin, Wai-Fong; Sam, Choon-Kook; Koh, Chong-Lek (2010). "Rapid degradation of N-3-oxo-acylhomoserine lactones by a Bacilluscereus isolate from Malaysian rainforest soil". Antonie van Leeuwenhoek. 98 (3): 299–305. doi:10.1007/s10482-010-9438-0. PMID 20376561.
- ↑ Franks, N.R.; Dornhaus, A.; et al. (2006). "Not everything that counts can be counted: ants use multiple metrics for a single nest trait". Proceedings of the Royal Society B: Biological Sciences. 273 (1583): 165–9. doi:10.1098/rspb.2005.3312. PMC 1560019
 . PMID 16555783.
. PMID 16555783. - ↑ Franks, N.R.; Hooper, J.; et al. (2005). "Tomb evaders: house-hunting hygiene in ants". Biology Letters. 1 (2): 190–2. doi:10.1098/rsbl.2005.0302. PMC 1626204
 . PMID 17148163.
. PMID 17148163. - ↑ Mallon, E.B.; Pratt, S.C.; et al. (2001). "Individual and collective decision-making during nest site selection by the ant Leptothorax albipennis". Behavioral Ecology and Sociobiology. 50 (4): 352–9. doi:10.1007/s002650100377.
- ↑ Pratt, S.C. (2005). "Quorum sensing by encounter rates in the ant Temnothorax albipennis". Behavioral Ecology. 16 (2): 488–96. doi:10.1093/beheco/ari020.
- ↑ Seeley, T.D.; Visscher, P.K. (2004). "Group decision making in nest-site selection by honey bees". Apidologie. 35 (2): 101–16. doi:10.1051/apido:2004004.
- ↑ Seeley, T.D.; Visscher, P.K. (2006). "Group decision making in honey bee swarms". American Scientist. 94 (3): 220–9. doi:10.1511/2006.3.220.
- ↑ Britton, M.; Sacks, L. (2004). "The SECOAS Project—Development of a Self-Organising, Wireless Sensor Network for Environmental Monitoring" (PDF). SANPA.
- ↑ Peysakhov, M.; Regli, W. (2005). "Ant inspired server population management in a service based computing environment". Proceedings 2005 IEEE Swarm Intelligence Symposium, 2005. SIS 2005. Swarm Intelligence Symposium, Proceedings 2005 IEEE. pp. 357–64. doi:10.1109/SIS.2005.1501643. ISBN 0-7803-8916-6.
- ↑ Sahin, E.; Franks, N. (2002). "Measurement of Space: From Ants to Robots". Proceedings of WGW 2002: EPSRC/BBSRC International Workshop. CiteSeerX 10.1.1.161.6407
 .
.
Further reading
- Dedicated issue of Philosophical Transactions B on quorum sensing. Some articles are freely available.
External links
- The Quorum Sensing Website
- Cell-to-Cell Communication in Bacteria
- The SECOAS project—Development of a Self-Organising, Wireless Sensor Network for Environmental Monitoring
- Measurement of Space: From Ants to Robots
- Instant insight into quorum sensing from the Royal Society of Chemistry
- Bonnie Bassler: Discovering bacteria's amazing communication system
- Bonnie Bassler's seminar: "Cell-Cell Communication in Bacteria"
