Oxysterol-binding protein
| Oxysterol-binding protein | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystallographic structure of the oxysterol-binding protein (rainbow color cartoon, N-terminus = blue, C-terminus = red) bound to 7-hydroxycholesterol (stick diagram, carbon = white, oxygen = red).[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Oxysterol_BP | ||||||||
| Pfam | PF01237 | ||||||||
| InterPro | IPR000648 | ||||||||
| PROSITE | PDOC00774 | ||||||||
| OPM superfamily | 173 | ||||||||
| OPM protein | 1zi7 | ||||||||
| |||||||||
The oxysterol-binding protein (OSBP)-related proteins (ORPs) are a family of lipid transfer proteins (LTPs). Concretely, they constitute a family of sterol and phosphoinositide binding and transfer proteins in eukaryotes[2] that are conserved from yeast to humans. They are lipid-binding proteins implicated in many cellular processes related with oxysterol, including signaling, vesicular trafficking, lipid metabolism, and nonvesicular sterol transport.
In yeast cells, it is probable that ORPs function as sterol transporters, perhaps in regions where organelle membranes are closely apposed. Various ORPs confine at membrane contacts sites (MCS), where endoplasmic reticulum (ER) is apposed with other organelle limiting membranes. Yeast ORPs also participate in vesicular trafficking, although their role is unclear.
In mammalian cells, some ORPs function as sterol sensors that regulate the assembly of protein complexes in response to changes in cholesterol levels.[3] By that means, ORPs most likely affect organelle membrane lipid compositions, with impacts on signaling and vesicle transport, but also cellular lipid metabolism. [4]
Oxysterol is a cholesterol metabolite that can be produced through enzymatic or radical processes. Oxysterols, that are the 27-carbon products of cholesterol oxidation by both enzymic and non-enzymic mechanisms, constitute a large family of lipids involved in a plethora of physiological processes. Studies identifying the specific cellular targets of oxysterol indicate that several oxysterols may be regulators of cellular lipid metabolism via control of gene transcription. In addition, they were shown to be involved in other processes such as immune regulatory functions and brain homeostasis.[5] [6]
Structure
.png)
All oxysterol related proteins (ORP) contain a core lipid-binding domain (ORD), which has a characteristic amino acids sequence, EQVSHHPP. The most studied ORP are human and yeast ones, and the only OSBP-ORP whose structure is completely known is the Kes1p, also called Osh4p, a yeast one. Six different protein domains and structural motifs types are found in OSBP-ORPs.[7]
.png)

FFAT motif
This is two phenylalanines in an acidic tract. It is bound by the endoplasmic reticulum to a lot of proteins involved in lipid metabolism. It is contained in most mammalian ORPs and in about 40% of yeast's ORPs.
Ankyrin motif
It is thought that it takes part in protein-protein interactions, but it is not known for certain. In some proteins, it also contributes to the localization of each protein to a membrane contact site (zone of close contact between the endoplasmic reticulum and a second organelle).
Transmembrane domain
It is only present in some human proteins. It is an hydrophobic region which holds the protein to the cell membrane.
PH (pleckstrin homology) domain
It binds phosphoinositides, usually only the ones which have low affinity and other ligands. It also recognizes organelles enriched in the PIPs.
GOLD (Golgi dynamics) domain
As well as Ankyrin motif, it probably mediates interactions between proteins. It is only found in one yeast protein and it is not found in any human ORP.
ORD (OSBP-related domain)
It contains the EQVSHHPP sequence. It has an hydrophobic pocket that binds a sterol and also contains multiple membrane binding surfaces which permit the protein to have the ability to cause liposome aggregation.
Principal functions
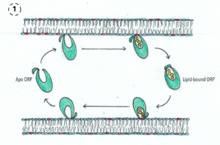
As part of the Lipid Transfer proteins (LTPs) family ,ORPs have different and variate functions. This functions include signaling, vesicular trafficking, lipid metabolism and nonvesicular sterol transport.[8] ORPs have been studied in many organisms cells as human cells or yeast. In yeast, where organelle membranes are closely apposed ORPs work as sterol transporters. They are also part of vesicular trafficking but their role is not clear yet. In mamalian, ORPs participate as sterol sensors.This sensors regulate the assembly of protein complexes when cholesterol levels fluctuate.[9]




They use the following mechanisms:
1-They could extract and deliver lipids from one membrane to another. Probably at membrane contact site.
2-ORPs help establish the membrane when transient changes in the distribution of lipids occur. They add or remove lipids within different regions of the membrane. The exclusion of certain lipids in particular regions drive to processes such as membrane binding or signaling.
3-They work as lipid sensors altering interactions with other proteins due to binding or releasing lipid ligands. It occurs mainly at inally organelle contact sites.
4-The access of other lipid-binding proteins to the membrane is regulated by ORPs in two ways. One way is by presenting a lipid to a second lipid-binding protein. (5)Another way is preventing the lipid-binding protein from accessing a lipid in the membrane. This two mechanisms are not mutually exclusive so ORPs might use both.
OSBP-ORPs human proteins
In humans there are 12 ORP genes, and splicing generates 16 different protein products.[10] [11] [12]
| Symbol | Name | Length (aa) | Chromosome | Molecular function |
|---|---|---|---|---|
| OSBP [13]
(OSBP1) |
Oxysterol-binding protein 1 | 807 | 11q12.1 |
|
| OSBP2 [14]
(KIAA1664, ORP-4, ORP4) |
Oxysterol-binding protein 2 | 916 | 22q12.2 |
|
| OSBPL1A [15]
(ORP-1, ORP1) |
Oxysterol-binding protein-related protein 1 | 950 | 18q11.2 |
|
| OSBPL2 [16]
"(KIAA0772, ORP2)" [17] |
Oxysterol-binding protein-related protein 2 | 480 | 20q13.33 |
|
| OSBPL3 [18]
(ORP-3, ORP3, KIAA0704) |
Oxysterol-binding protein-related protein 3 | 887 | 7p15.3 |
|
| OSBPL5 [19]
(KIAA1534, ORP5) |
Oxysterol-binding protein-related protein 5 | 879 | 11p15.4 |
|
| OSBPL6 [20]
(ORP6) |
Oxysterol-binding protein-related protein 6 | 934 | 2q31.2 |
|
| OSBPL7 [21]
(ORP7, MGC71150) |
Oxysterol-binding protein-related protein 7 | 842 | 17q21 |
|
| OSBPL8 [22]
(OSBP10, ORP8, MST120, MSTP120) |
Oxysterol-binding protein-related protein 8 | 889 | 12q14 |
|
| OSBPL9 [23]
(ORP9, OSBP4) |
Oxysterol-binding protein-related protein 9 | 736 | 1p32.3 |
|
| OSBPL10 [24]
(ORP10, OSBP9) |
Oxysterol-binding protein-related protein 10 | 764 | 3p23 |
|
| OSBPL11 [25]
(ORP-11, ORP11, FLJ13012, FLJ13164) |
Oxysterol-binding protein-related protein 11 | 747 | 3q21.2 |
|
OSBP-ORPs yeast proteins
In yeast (Saccharomyces cerevisiæ) we can find 7 ORP genes called OSH1-7, but they have some additional names as well.[26] [27]
| Symbol | Name | Length (aa) | Molecular function | Subcellular location |
|---|---|---|---|---|
| OSH1 [28]
(SWH1, YAR042W, YAR044W) |
Oxysterol-binding protein homolog 1 | 1188 |
|
|
| OSH2 [29]
(YDL019C, D2845) |
Oxysterol-binding protein homolog 2 | 1283 |
|
|
| OSH3 [30]
(YHR073W) |
Oxysterol-binding protein homolog 3 | 996 |
|
|
| OSH4 [31]
(KES1, YPL145C, LPI3C, P2614) |
Oxysterol-binding protein homolog 4 | 434 |
|
|
| OSH5 [32]
(HES1, YOR237W, O5234) |
Oxysterol-binding protein homolog 5 | 434 |
|
- |
| OSH6 [33]
(YKR003W, YK102) |
Oxysterol-binding protein homolog 6 | 448 |
|
|
| OSH7 [34]
(YHR001W) |
Oxysterol-binding protein homolog 7 | 437 |
|
|
Curiosities
Some oxysterols have been found to contribute to the inflammation and oxidative damage as well as in cell death in the appearance and especially the development of some of the most important chronic diseases, such as atherosclerosis, neurodegenerative diseases, inflammatory bowel diseases, age-related macular degeneration and other pathological conditions related to cholesterol absorption.[35]
Besides, a recent study suggests a method of screening and diagnosing Niemann-Pick C disease by plasma oxysterol screening, which is found to be less invasive, more sensitive and specific and more economical strategy than the current practice.[36]
References
- ↑ Im, Y. J.; Raychaudhuri, S.; Prinz, W. A.; Hurley, J. H. (2005). "Structural mechanism for sterol sensing and transport by OSBP-related proteins". Nature. 437 (7055): 154–158. doi:10.1038/nature03923. PMC 1431608
 . PMID 16136145. PDB: 1ZHT; Im, Y. J.; Raychaudhuri, S.; Prinz, W. A.; Hurley, J. H. (2005). "Structure of yeast oxysterol binding protein Osh4 in complex with 7-hydroxycholesterol". doi:10.2210/pdb1zht/pdb.
. PMID 16136145. PDB: 1ZHT; Im, Y. J.; Raychaudhuri, S.; Prinz, W. A.; Hurley, J. H. (2005). "Structure of yeast oxysterol binding protein Osh4 in complex with 7-hydroxycholesterol". doi:10.2210/pdb1zht/pdb. - ↑ Weber-Boyvat, Marion; Zhong, Wenbin; Yan, Daoguang; Olkkonen, Vesa M. (1 July 2013). "Oxysterol-binding proteins: functions in cell regulation beyond lipid metabolism". Biochemical Pharmacology. pp. 89–95. doi:10.1016/j.bcp.2013.02.016.
- ↑ Raychaudhuri, Sumana; Prinz, William A. (10 November 2010). "The Diverse Functions of Oxysterol-Binding Proteins". Annual review of cell and developmental biology. pp. 157–177. doi:10.1146/annurev.cellbio.042308.113334.
- ↑ Weber-Boyvat, Marion; Zhong, Wenbin; Yan, Daoguang; Olkkonen, Vesa M. (1 July 2013). "Oxysterol-binding proteins: functions in cell regulation beyond lipid metabolism". Biochemical Pharmacology. pp. 89–95. doi:10.1016/j.bcp.2013.02.016.
- ↑ Mutemberezi, Valentin; Guillemot-Legris, Owein; Muccioli, Giulio G. (26 September 2016). "Oxysterols: From cholesterol metabolites to key mediators". Progress in Lipid Research. pp. 152–169. doi:10.1016/j.plipres.2016.09.002.
- ↑ van Reyk, David M.; Brown, Andrew J.; Hult'en, Lillemor Mattsson; Dean, Roger T.; Jessup, Wendy (1 January 2006). "Oxysterols in biological systems: sources, metabolism and pathophysiological relevance". Redox Report: Communications in Free Radical Research. pp. 255–262. doi:10.1179/135100006X155003.
- ↑ Raychaudhuri, Sumana; Prinz, William A. (10 November 2010). "The Diverse Functions of Oxysterol-Binding Proteins". Annual review of cell and developmental biology. pp. 157–177. doi:10.1146/annurev.cellbio.042308.113334.
- ↑ Ngo, Mike; Ridgway, Neale D. (1 March 2009). "Oxysterol binding protein-related Protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function". Molecular Biology of the Cell. pp. 1388–1399. doi:10.1091/mbc.E08-09-0905.
- ↑ Raychaudhuri, Sumana; Prinz, William A. (10 November 2010). "The Diverse Functions of Oxysterol-Binding Proteins". Annual review of cell and developmental biology. pp. 157–177. doi:10.1146/annurev.cellbio.042308.113334.
- ↑ Lehto, M.; Laitinen, S.; Chinetti, G.; Johansson, M.; Ehnholm, C.; Staels, B.; Ikonen, E.; Olkkonen, V. M. (1 August 2001). "The OSBP-related protein family in humans". Journal of Lipid Research. pp. 1203–1213.
- ↑ "Oxysterol binding proteins (OSBP) Gene Family | HUGO Gene Nomenclature Committee". www.genenames.org. Retrieved 11 October 2016.
- ↑ ""oxysterol binding" proteins in UniProtKB". www.uniprot.org. Retrieved 11 October 2016.
- ↑ "OSBP - Oxysterol-binding protein 1 - Homo sapiens (Human) - OSBP gene & protein". www.uniprot.org.
- ↑ "OSBP2 - Oxysterol-binding protein 2 - Homo sapiens (Human) - OSBP2 gene & protein". www.uniprot.org.
- ↑ "OSBPL1A - Oxysterol-binding protein-related protein 1 - Homo sapiens (Human) - OSBPL1A gene & protein". www.uniprot.org.
- ↑ "OSBPL2 - Oxysterol-binding protein-related protein 2 - Homo sapiens (Human) - OSBPL2 gene & protein". www.uniprot.org.
- ↑ Escajadillo, Tamara; Wang, Hongxia; Li, Linda; Li, Donghui; Sewer, Marion B. (15 May 2016). "Oxysterol-related-binding-protein related Protein-2 (ORP2) regulates cortisol biosynthesis and cholesterol homeostasis". Molecular and Cellular Endocrinology. pp. 73–85. doi:10.1016/j.mce.2016.03.006.
- ↑ "OSBPL3 - Oxysterol-binding protein-related protein 3 - Homo sapiens (Human) - OSBPL3 gene & protein". www.uniprot.org.
- ↑ "OSBPL5 - Oxysterol-binding protein-related protein 5 - Homo sapiens (Human) - OSBPL5 gene & protein". www.uniprot.org.
- ↑ "OSBPL6 - Oxysterol-binding protein-related protein 6 - Homo sapiens (Human) - OSBPL6 gene & protein". www.uniprot.org.
- ↑ "OSBPL7 - Oxysterol-binding protein-related protein 7 - Homo sapiens (Human) - OSBPL7 gene & protein". www.uniprot.org.
- ↑ "OSBPL8 - Oxysterol-binding protein-related protein 8 - Homo sapiens (Human) - OSBPL8 gene & protein". www.uniprot.org.
- ↑ "OSBPL9 - Oxysterol-binding protein-related protein 9 - Homo sapiens (Human) - OSBPL9 gene & protein". www.uniprot.org.
- ↑ "OSBPL10 - Oxysterol-binding protein-related protein 10 - Homo sapiens (Human) - OSBPL10 gene & protein". www.uniprot.org.
- ↑ "OSBPL11 - Oxysterol-binding protein-related protein 11 - Homo sapiens (Human) - OSBPL11 gene & protein". www.uniprot.org.
- ↑ Beh, C. T.; Cool, L.; Phillips, J.; Rine, J.; Ehnholm, C.; Staels, B.; Ikonen, E.; Olkkonen, V. M. (1 March 2001). "Overlapping functions of the yeast oxysterol-binding protein homologues". Genetics. pp. 1117–1140.
- ↑ ""oxysterol binding" proteins in UniProtKB". www.uniprot.org. Retrieved 17 October 2016.
- ↑ "SWH1 - Oxysterol-binding protein homolog 1 - Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) - SWH1 gene & protein". www.uniprot.org.
- ↑ "OSH2 - Oxysterol-binding protein homolog 2 - Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) - OSH2 gene & protein". www.uniprot.org.
- ↑ "OSH3 - Oxysterol-binding protein homolog 3 - Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) - OSH3 gene & protein". www.uniprot.org.
- ↑ "KES1 - Oxysterol-binding protein homolog 4 - Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) - KES1 gene & protein". www.uniprot.org.
- ↑ "HES1 - Protein HES1 - Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) - HES1 gene & protein". www.uniprot.org.
- ↑ "OSH6 - Oxysterol-binding protein homolog 6 - Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) - OSH6 gene & protein". www.uniprot.org.
- ↑ "OSH7 - Oxysterol-binding protein homolog 7 - Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) - OSH7 gene & protein". www.uniprot.org.
- ↑ Poli, Giuseppe; Biasi, Fiorella; Leonarduzzi, Gabriella (31 January 2013). "Oxysterols in the pathogenesis of major chronic diseases". Redox Biology. pp. 125–130. doi:10.1016/j.redox.2012.12.001.
- ↑ van Karnebeek, Clara D.M.; Mohammadi, Tima; Tsao, Nicole; Sinclair, Graham; Sirrs, Sandra; Stockler, Sylvia; Marra, Carlo (1 July 2014). "Health economic evaluation of plasma oxysterol screening in the diagnosis of Niemann–Pick Type C disease among intellectually disabled using discrete event simulation". Molecular Genetics and Metabolism. pp. 226–232. doi:10.1016/j.ymgme.2014.07.004.
This article incorporates text from the public domain Pfam and InterPro IPR000648