Oocyte
| Oocyte | |
|---|---|
| Identifiers | |
| MeSH | A05.360.490.690.680 |
An oocyte (UK /ˈoʊəsʌɪt/, US /ˈoʊ.oʊ.saɪt/), oöcyte, ovocyte, or rarely ocyte, is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in the ovary during female gametogenesis. The female germ cells produce a primordial germ cell (PGC), which then undergoes mitosis, forming oogonia. During oogenesis, the oogonia become primary oocytes. An oocyte is a form of genetic material that can be collected for cryoconservation. Cryoconservation of animal genetic resources have been put into action as a means of conserving traditional livestock.
Formation
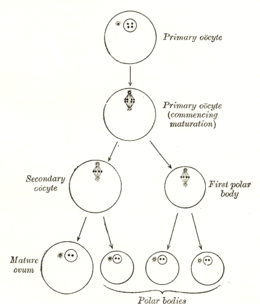
The formation of an oocyte is called oocytogenesis, which is a part of oogenesis.[1] Oogenesis results in the formation of both primary oocytes during fetal period, and of secondary oocytes after it as part of ovulation.
| Cell type | ploidy/chromosomes | chromatids | Process | Time of completion |
| Oogonium | diploid/46(2N) | 2C | Oocytogenesis (mitosis) | third trimester |
| primary Oocyte | diploid/46(2N) | 4C | Ootidogenesis (meiosis I) (Folliculogenesis) | Dictyate in prophase I for up to 50 years |
| secondary Oocyte | haploid/23(1N) | 2C | Ootidogenesis (meiosis II) | Halted in metaphase II until fertilization |
| Ootid | haploid/23(1N) | 1C | Ootidogenesis (meiosis II) | Minutes after fertilization |
| Ovum | haploid/23(1N) | 1C |
Characteristics
Cytoplasm
Oocytes are rich in cytoplasm, which contains yolk granules to nourish the cell early in development.
Nucleus
During the primary oocyte stage of oogenesis, the nucleus is called a germinal vesicle.[2]
The only normal human type of secondary oocyte has the 23rd (sex) chromosome as 23,X (female-determining), whereas sperm can have 23,X (female-determining) or 23,Y (male-determining).
Nest
The space within an ovum or immature ovum is located is the cell-nest.[3]
Maternal contributions
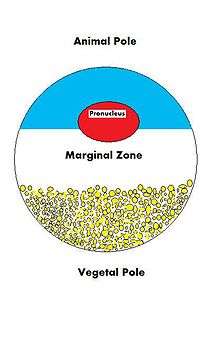
Because the fate of an oocyte is to become fertilized and ultimately grow into a fully functioning organism, it must be ready to regulate multiple cellular and developmental processes. The oocyte, a large and complex cell, must be supplied with numerous molecules that will direct the growth of the embryo and control cellular activities. As the oocyte is a product of female gametogenesis, the maternal contribution to the oocyte and consequently the newly fertilized egg is enormous. There are many types of molecules that are maternally supplied to the oocyte, which will direct various activities within the growing zygote.
Avoidance of damage to germ-line DNA
The DNA of a cell is vulnerable to the damaging effect of oxidative free radicals produced as byproducts of cellular metabolism. DNA damage occurring in oocytes, if not repaired, can be lethal and result in reduced fecundity and loss of potential progeny. Oocytes are substantially larger than the average somatic cell, and thus considerable metabolic activity is necessary for their provisioning. If this metabolic activity were carried out by the oocyte’s own metabolic machinery, the oocyte genome would be exposed to the reactive oxidative by-products generated. Thus it appears that a process evolved to avoid this vulnerability of germ line DNA. It was proposed that, in order to avoid damage to the DNA genome of the oocytes, the metabolism contributing to the synthesis of much of the oocyte’s constituents was shifted to other maternal cells that then transferred these constituents to oocytes.[4][5] Thus, oocytes of many organisms are protected from oxidative DNA damage while storing up a large mass of substances to nurture the zygote in its initial embryonic growth.
mRNAs and proteins
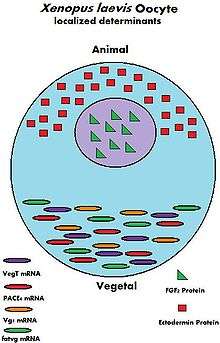
During the growth of the oocyte, a variety of maternally transcribed messenger RNAs, or mRNAs, are supplied by maternal cells. These mRNAs can be stored in mRNP (message ribonucleoprotein) complexes and be translated at specific time points, they can be localized within a specific region of the cytoplasm, or they can be homogeneously dispersed within the cytoplasm of the entire oocyte.[6] Maternally loaded proteins can also be localized or ubiquitous throughout the cytoplasm. The translated products of the mRNAs and the loaded proteins have multiple functions; from regulation of cellular "house-keeping" such as cell cycle progression and cellular metabolism, to regulation of developmental processes such as fertilization, activation of zygotic transcription, and formation of body axes.[6] Below are some examples of maternally inherited mRNAs and proteins found in Xenopus laevis oocytes.
| Name | Type of maternal molecule | Localization | Function |
|---|---|---|---|
| VegT[7] | mRNA | Vegetal hemisphere | Transcription factor |
| Vg1[8] | mRNA | Vegetal hemisphere | Transcription factor |
| XXBP-1[9] | mRNA | Not known | Transcription factor |
| CREB[10] | Protein | Ubiquitous | Transcription factor |
| FoxH1[11] | mRNA | Ubiquitous | Transcription factor |
| p53[12] | Protein | Ubiquitous | Transcription Factor |
| Lef/Tcf[13] | mRNA | Ubiquitous | Transcription factor |
| FGF2[14] | Protein | Nucleus | Not known |
| FGF2, 4, 9 FGFR1[13] | mRNA | Not known | FGF signaling |
| Ectodermin[15] | Protein | Animal hemisphere | Ubiquitin ligase |
| PACE4[16] | mRNA | Vegetal hemisphere | Proprotein convertase |
| Coco[17] | Protein | Not known | BMP inhibitor |
| Twisted gastrulation[13] | Protein | Not known | BMP/Chordin binding protein |
| fatvg[18] | mRNA | Vegetal hemisphere | Germ cell formation and cortical rotation |
Mitochondria
The oocyte receives mitochondria from maternal cells, which will go on to control embryonic metabolism and apoptotic events.[6] The partitioning of mitochondria is carried out by a system of microtubules that will localize mitochondria throughout the oocyte. In certain organisms, such as mammals, paternal mitochondria brought to the oocyte by the spermatozoon are degraded through the attachment of ubiquitinated proteins. The destruction of paternal mitochondria ensures the strictly maternal inheritance of mitochondria and mitochondrial DNA or mtDNA.[6]
Nucleolus
In mammals, the nucleolus of the oocyte is derived solely from maternal cells.[19] The nucleolus, a structure found within the nucleus, is the location where rRNA is transcribed and assembled into ribosomes. While the nucleolus is dense and inactive in a mature oocyte, it is required for proper development of the embryo.[19]
Ribosomes
Maternal cells also synthesize and contribute a store of ribosomes that are required for the translation of proteins before the zygotic genome is activated. In mammalian oocytes, maternally derived ribosomes and some mRNAs are stored in a structure called cytoplasmic lattices. These cytoplasmic lattices, a network of fibrils, protein, and RNAs, have been observed to increase in density as the number of ribosomes decrease within a growing oocyte.[20]
Paternal contributions
The spermatozoon that fertilizes an oocyte will contribute its pronucleus, the other half of the zygotic genome. In some species, the spermatozoon will also contribute a centriole, which will help make up the zygotic centrosome required for the first division. However, in some species, such as in the mouse, the entire centrosome is acquired maternally.[21] Currently under investigation is the possibility of other cytoplasmic contributions made to the embryo by the spermatozoon.
During fertilization, the sperm provides three essential parts to the oocyte: (1) a signalling or activating factor, which causes the metabolically dormant oocyte to activate; (2) the haploid paternal genome; (3) the centrosome, which is responsible for maintaining the microtubule system. See anatomy of sperm
Abnormalities
- nondisjunction—a failure of proper homolog separation in meiosis I, or sister chromatid separation in meiosis II can lead to aneuploidy, in which the oocyte has the wrong number of chromosomes, for example 22,X or 24,X. This is the cause of conditions like Down syndrome and Edwards syndrome in humans. It is more likely with advanced maternal age.
- Some oocytes have multiple nuclei, although it is thought they never mature.
See also
- Cortical granule
- Cryoconservation of animal genetic resources
- Folliculogenesis
- Oocyte maturation inhibitor
- Polar body
- Symmetry breaking and cortical rotation
References
- ↑ answers.com
- ↑ Biology-online
- ↑ Grier HJ, Uribe MC, Parenti LR (April 2007). "Germinal epithelium, folliculogenesis, and postovulatory follicles in ovaries of rainbow trout, Oncorhynchus mykiss (Walbaum, 1792) (Teleostei, protacanthopterygii, salmoniformes)". J. Morphol. 268 (4): 293–310. doi:10.1002/jmor.10518. PMID 17309079.
- ↑ Bernstein C. (1993). Sex as a response to oxidative DNA damage. Chapter 10 (see pages 204-205) in “DNA and Free Radicals” (editors: Barry Halliwell, Okezie I Aruoma). Ellis Horwood Limited (publisher), Great Gritain ISBN 0-13-222035-0
- ↑ Bernstein, C. (1998). Sex as a response to oxidative DNA damage. Chapter 4, see pages 112-113. In “DNA and Free Radicals: Techniques, Mecchanisms & Applications” (editors: Okezie I Aruoma, Barry Halliwell). OICA International (publisher), Saint Lucia and London ISBN 976-8056169
- 1 2 3 4 Mtango N.R., Potireddy S., Latham K.E. (2008). Oocyte quality and maternal control of development. Int. Rev. Cell Mol. Biol. 268, 223-290.
- ↑ Zhang J., King M.L. (1996). Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development. 12, 4119–29.
- ↑ Heasman J., Wessely O., Langland R., Craig E.J., Kessler D.S. (2001). Vegetal localization of maternal mRNAs is disrupted by VegT depletion. Dev Biol. 240, 377–386.
- ↑ Zhao H., Cao Y., Grunz H. (2003). Xenopus X-box binding protein 1, a leucine zipper transcription factor, is involved in the BMP signaling pathway. Dev Biol. 257, 278–291.
- ↑ Sundaram N., Tao Q., Wylie C., Heasman J. (2003). The role of maternal CREB in early embryogenesis of Xenopus laevis. Dev Biol. 261, 337–352.
- ↑ Kofron M., Puck H., Standley H., Wylie C., Old R., Whitman M., et al. (2004). New roles for FoxH1 in patterning the early embryo. Development. 131, 5065–5078.
- ↑ Takebayashi-Suzuki K., Funami J., Tokumori D., Saito A., Watabe T., Miyazono K., et al. (2003). Interplay between the tumor suppressor p53 and TGF beta signaling shapes embryonic body axes in Xenopus. Development. 130, 3929–3939.
- 1 2 3 Heasman, J. (2006). Maternal determinants of embryonic cell fate. Semin. Cell Dev. Biol. 17, 93-98.
- ↑ Song J., Slack J.M. (1994). Spatial and temporal expression of basic fibroblast growth factor (FGF-2) mRNA and protein in early Xenopus development. Mech Dev. 48, 141–151.
- ↑ Dupont S., Zacchigna L., Cordenonsi M., Soligo S., Adorno M., Rugge M., et al. (2005). Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 121, 87–99.
- ↑ Birsoy B., Berg L., Williams P.H., Smith J.C., Wylie C.C., Christian J.L., et al. (2005). XPACE4 is a localized pro-protein convertase required for mesoderm induction and the cleavage of specific TGFbeta proteins in Xenopus development. Development. 132, 591–602.
- ↑ Bell E., Munoz-Sanjuan I., Altmann C.R., Vonica A., Brivanlou A.H. (2003). Cell fate specification and competence by Coco, a maternal BMP, TGFbeta and Wnt inhibitor. Development. 130, 1381–1389.
- ↑ Chan A. P., Kloc M., Larabell C. A., LeGros M., Etkin L.D. (2007). The maternally localized RNA fatvg is required for cortical rotation and germ cell formation. Mech Dev. 124, 350-363.
- 1 2 Ogushi S., et al. (2008). The maternal nucleolus is essential for early embryonic development in mammals. Science. 319, 613-616
- ↑ Vitale A.M., Yurttas P., Fitzhenry R.J., Cohen-Gould, L., Wu W., Gossen J.A., Coonrod S.A. (2009). Role for PADI6 and the CPLs in ribosomal storage in oocytes and translation in the early embryo. Development. 135, 2627-2636.
- ↑ Sutovsky P., Schatten G. (2000). Paternal contributions to the mammalian zygote: fertilization after sperm-egg fusion. Int. Rev. Cytol. 195, 1-65.
Sources
- William K. Purves, Gordon H. Orians, David Sadava, H. Craig Heller, Craig Heller (2003). Life: The Science of Biology (7th ed.), pp. 823–824.
External links
| Preceded by None |
Stages of human development Sperm + Oocyte |
Succeeded by Zygote |