Mesomycetozoea
| Mesomycetozoea | |
|---|---|
 | |
| Scientific classification | |
| Domain: | Eukarya |
| (unranked) | Opisthokonta |
| (unranked) | Choanozoa or Holozoa |
| Class: | Mesomycetozoea Mendoza et al. 2002 |
| Orders | |
| Synonyms | |
|
Ichthyosporea Cavalier-Smith 1998[1] | |
The Mesomycetozoea (or DRIP clade, or Ichthyosporea) are a small group of Opisthokonta in Eukarya (formerly protists), mostly parasites of fish and other animals.
Significance
They are not particularly distinctive morphologically, appearing in host tissues as enlarged spheres or ovals containing spores, and most were originally classified in various groups as fungi, protozoa, or colorless algae. However, they form a coherent group on molecular trees, closely related to both animals and fungi and so of interest to biologists studying their origins. In a 2008 study they emerge robustly as the sister-group of the clade Filozoa, which includes the animals.[2]
Huldtgren et al., following x-ray tomography of microfossils of the Ediacaran Doushantuo Formation, has interpreted them as mesomycetozoan spore capsules.[3]
Terminology
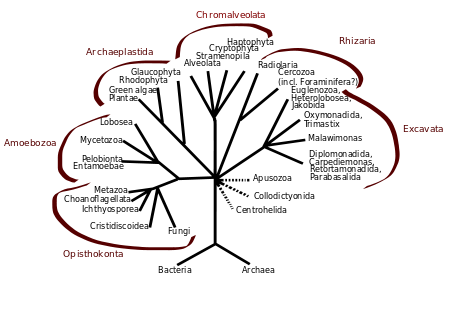
The name DRIP is an acronym for the first protozoa identified as members of the group,[4] Cavalier-Smith later treated them as the class Ichthyosporea, since they were all parasites of fish.
- order Dermocystida
- "D": Dermocystidium. One species, Rhinosporidium seeberi, infects birds and mammals, including humans.
- "R": the "rosette agent", now known as Sphaerothecum destruens
- order Ichthyophonida
- "I": Ichthyophonus
- "P": Psorospermium
Since other new members have been added (e.g. the former fungal orders Eccrinales and Amoebidiales), Mendoza et al. suggested changing the name to Mesomycetozoea, which refers to their evolutionary position. On Eukaryota tree, in Opisthokont clade, Mesomycetozoea is in the middle ("Meso-") of the fungi ("-myceto-") and the animals ("-zoea").[5] Note the name Mesomycetozoa (without a second e) is also used to refer to this group, but Mendoza et al. use it as an alternate name for the phylum Choanozoa.[6]
Taxonomy
- Class Ichthyosporea Cavalier-Smith 1998[7][8]
- Order Dermocystida Cavalier-Smith 1998
- Family Rhinosporidiaceae Mendoza et al. 2001
- Genus Sphaerothecum Arkush et al. 2003 (Rosette agent)
- Genus Rhinosporidium Minchin & Fantham 1905
- Genus Amphibiocystidium Pascolini et al. 2003
- Genus Dermocystidium Pérez 1908 [Dermocystis Pérez 1907 non]
- Genus Amphibiothecum Feldman, Wimsatt & Green, 2005
- Genus Dermosporidium Carini 1940
- Family Rhinosporidiaceae Mendoza et al. 2001
- Order Ichthyophonida Cavalier-Smith 1998
- Suborder Sphaeroformina Cavalier-Smith 2012
- Genus Caullerya Chatton 1907
- Family Psorospermidae Cavalier-Smith 2012
- Genus Psorospermis Cavalier-Smith 2012 [Psorospermium Hilgendorf 1883 non Eimer 1870]
- Family Piridae Cavalier-Smith 2012
- Genus Abeoforma Marshall & Berbee 2011
- Genus Pirum Marshall & Berbee 2011
- Family Creolimacidae Cavalier-Smith 2012
- Genus Anurofeca Baker, Beebee & Ragan 1999
- Genus Pseudoperkinsus Figueras et al. 2000
- Genus Creolimax Marschall et al. 2008
- Genus Sphaeroforma Jostensen et al. 2002
- Suborder Trichomycina Cavalier-Smith 2012
- Family Ichthyophonidae Cavalier-Smith 2012
- Genus Ichthyophonus Plehn & Mulsow 1911
- Family Amoebidiidae Lichtenstein 1917 ex Kirk et al. 2001
- Genus Amoebidium Cienkowski 1861
- Genus Paramoebidium Léger & Duboscq 1929
- Family Palavasciaceae Manier & Lichtward 1968
- Genus Palavascia Tuzet & Manier 1947 ex Lichtwardt 1964
- Family Parataeniellaceae Manier & Lichtward 1968
- Genus Lajassiella Tuzet & Manier 1951 ex Manier 1968
- Genus Nodocrinella Scheer 1977
- Genus Parataeniella Poiss. 1929
- Family Eccrinaceae Leger & Duboscq 1929
- Genus Alacrinella Manier & Ormières ex Manier 1968
- Genus Arundinula Léger & Duboscq 1906 [Arundinella Léger & Duboscq 1905 non Raddi 1823]
- Genus Astreptonema Hauptfleisch 1895 [Astereptonema Hauptfleisch 1895; Eccrinella Léger & Duboscq 1933]
- Genus Eccrinidus Manier 1970
- Genus Eccrinoides Léger & Duboscq 1929
- Genus Enterobryus [Andohaheloa Manier 1955; Capillus Granata 1908; Cestodella Tuzet, Manier & Jolivet 1957; Daloala Tuzet, Manier & Vog.-Zuber 1952; Eccrina Leidy 1852; Eccrinopsis Léger & Duboscq 1916; Lactella Maessen 1955; Paratrichella Manier 1947; Pistillaria Jeekel et al. 1959 non Fries 1821; Recticoma Scheer 1935; Trichella Léger & Duboscq 1929; Trichellopsis Maessen 1955]
- Genus Enteromyces Lichtwardt 1961
- Genus Enteropogon Hibbits 1979 non Nees 1836
- Genus Leidyomyces Lichtwardt et al. 1999
- Genus Paramacrinella Manier & Grizel 1971
- Genus Passalomyces Lichtwardt et al. 1999
- Genus Ramacrinella Manier & Ormiéres 1962 ex Manier 1968
- Genus Taeniella Léger & Duboscq 1911
- Genus Taeniellopsis Poiss. 1927
- Family Ichthyophonidae Cavalier-Smith 2012
- Suborder Sphaeroformina Cavalier-Smith 2012
- Order Dermocystida Cavalier-Smith 1998
References
- ↑ Cavalier-Smith, T. 1998. Neomonada and the origin of animals and fungi. In: Coombs GH, Vickerman K, Sleigh MA, Warren A (ed.) Evolutionary relationships among protozoa. Kluwer, London, pp. 375-407,
- ↑ Shalchian-Tabrizi K., Minge M.A., Espelund M.; et al. (7 May 2008). Aramayo, Rodolfo, ed. "Multigene Phylogeny of Choanozoa and the Origin of Animals". PLoS ONE. 3 (5): e2098. doi:10.1371/journal.pone.0002098. PMC 2346548
 . PMID 18461162.
. PMID 18461162. - ↑ Douglas Fox, "How life got complicated", Discover Magazine, December 2012.
- ↑ Ragan MA, Goggin CL, Cawthorn RJ, et al. (October 1996). "A novel clade of protistan parasites near the animal-fungal divergence". Proc. Natl. Acad. Sci. U.S.A. 93 (21): 11907–12. doi:10.1073/pnas.93.21.11907. PMC 38157
 . PMID 8876236.
. PMID 8876236. - ↑ Herr RA, Ajello L, Taylor JW, Arseculeratne SN, Mendoza L (September 1999). "Phylogenetic Analysis of Rhinosporidium seeberi's 18S Small-Subunit Ribosomal DNA Groups This Pathogen among Members of the Protoctistan Mesomycetozoa Clade". J. Clin. Microbiol. 37 (9): 2750–4. PMC 85368
 . PMID 10449446.
. PMID 10449446. - ↑ Mendoza L, Taylor JW, Ajello L (October 2002). "The class mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary". Annu. Rev. Microbiol. 56: 315–44. doi:10.1146/annurev.micro.56.012302.160950. PMID 12142489.
- ↑ Cavalier-Smith (May 2012). "Early evolution of eukaryote feeding modes, cell structural diversity, and classification of the protozoan phyla Loukozoa, Sulcozoa, and Choanozoa". European Journal of Protistology. 49: 115–178. doi:10.1016/j.ejop.2012.06.001.
- ↑ Crous PW, Gams W, Stalpers JA, Cannon PF, Kirk PM, David JC, Triebel D (November 2004). "An online database of names and descriptions as an alternative to registration". Mycological Research. 108 (11): 1236–1238. doi:10.1017/S0953756204221554.
- "Eccrinida". Species Fungorum. 2016.