Mechanistic target of rapamycin
| View/Edit Human | View/Edit Mouse |
The mechanistic target of rapamycin (mTOR), (formerly mammalian target of rapamycin before it was recognized to be highly conserved among eukaryotes.[4]) also known as FK506-binding protein 12-rapamycin-associated protein 1 (FRAP1), is a kinase that in humans is encoded by the MTOR gene.[5][6] mTOR is a member of the phosphatidylinositol 3-kinase-related kinase family of protein kinases.[7]
mTOR links with other proteins and serves as a core component of two distinct protein complexes, mTOR complex 1 and mTOR complex 2, which regulate different cellular processes.[8] In particular, as a core component of both complexes, mTOR functions as a serine/threonine protein kinase that regulates cell growth, cell proliferation, cell motility, cell survival, protein synthesis, autophagy, and transcription.[8][9] As a core component of mTORC2, mTOR also functions as a tyrosine protein kinase that promotes the activation of insulin receptors and insulin-like growth factor 1 receptors.[10] mTORC2 has also been implicated in the control and maintenance of the actin cytoskeleton.[8][11]
Discovery
mTOR was first named as the mammalian target of rapamycin. Rapamycin (sirolimus) was discovered in a soil sample from Easter Island, known locally as Rapa Nui, in the 1970s.[12] The bacterium Streptomyces hygroscopicus, isolated from that sample, produces an antifungal that researchers named rapamycin after the island.[13]
Rapamycin arrests fungal activity at the G1 phase of the cell cycle. In mammals, it suppresses the immune system by blocking the G1 to S phase transition in T-lymphocytes.[14] Thus, it is used as an immunosuppressant following organ transplantation.[15]
Molecular genetic studies in yeast (published in 1991) first identified FKBP12, TOR1, and TOR2 as the targets of rapamycin; these studies were conducted at the Biozentrum in Basel, Switzerland and Sandoz Pharmaceuticals (now Novartis) by Joseph Heitman, Rao Movva, and Michael N. Hall. They isolated rapamycin-resistant mutants of Saccharomyces cerevisiae and discovered that mutations in any of three genes can confer rapamycin resistance.[16] Two of the genes were named TOR1 and TOR2 for targets of rapamycin (TOR) and in honor of the Spalentor, a gate to the city of Basel where TOR was first discovered.[17] The third gene is FPR1, which encodes the yeast ortholog of FKBP12 binding protein in the TOR complexes. Loss of function mutations in FPR1 confer resistance to rapamycin, and also to FK506, providing genetic evidence the FKBP12-drug complexes are the active intracellular agents. Mutations in TOR1 or TOR2 that confer FKBP12-rapamycin resistance are dominant gain of function mutations that alter single amino acid residues within the FRB domain and thereby block FKBP12-rapamycin binding.[18] Several years later, in 1994 the mammalian target of rapamycin (mTOR) was identified and found to be the ortholog of the yeast Tor1/2 proteins and defined as the rapamycin target in mammals by David M. Sabatini and Solomon H. Snyder (Johns Hopkins University) and also by Robert Abraham (who first named it mTOR)[19] and Stuart L. Schreiber (Harvard University).[5] mTOR was named based on the precedent that TOR was first discovered via genetic and molecular studies of rapamycin-resistant mutants of Saccharomyces cerevisiae that identified Tor1 and Tor2 as the targets of rapamycin. Several groups also described the protein independently in the year 1994 using names such as FRAP (FKBP12–rapamycin-associated protein), RAFT1 (rapamycin and FKBP12 target 1), RAPT1 (rapamycin target 1) and SEP (sirolimus effector protein) to refer to the protein.[9][20] Due to the ubiquity of mTOR in animals the meaning of the m has been formally changed from "mammalian" to "mechanistic".
Function
MTOR integrates the input from upstream pathways, including insulin, growth factors (such as IGF-1 and IGF-2), and amino acids.[9] mTOR also senses cellular nutrient, oxygen, and energy levels.[21] The mTOR pathway is a central regulator of mammalian metabolism and physiology, with important roles in the function of tissues including liver, muscle, white and brown adipose tissue, and the brain, and is dysregulated in human diseases, such as diabetes, obesity, depression, and certain cancers.[22][23] Rapamycin inhibits mTOR by associating with its intracellular receptor FKBP12.[24][25] The FKBP12-rapamycin complex binds directly to the FKBP12-Rapamycin Binding (FRB) domain of mTOR, inhibiting its activity.[25]
Complexes
MTOR is the catalytic subunit of two structurally distinct complexes: mTORC1 and mTORC2.[26] Both complexes localize to different subcellular compartments, thus affecting their activation and function.[27]
mTORC1
mTOR Complex 1 (mTORC1) is composed of MTOR, regulatory-associated protein of MTOR (Raptor), mammalian lethal with SEC13 protein 8 (MLST8) and the non-core components PRAS40 and DEPTOR.[28][29] This complex functions as a nutrient/energy/redox sensor and controls protein synthesis.[9][28] The activity of mTORC1 is regulated by rapamycin, insulin, growth factors, phosphatidic acid, certain amino acids and their derivatives (e.g., L-leucine and β-hydroxy β-methylbutyric acid), mechanical stimuli, and oxidative stress.[28][30][31]
mTORC2
mTOR Complex 2 (mTORC2) is composed of MTOR, rapamycin-insensitive companion of MTOR (RICTOR), MLST8, and mammalian stress-activated protein kinase interacting protein 1 (mSIN1).[32][33] mTORC2 has been shown to function as an important regulator of the actin cytoskeleton through its stimulation of F-actin stress fibers, paxillin, RhoA, Rac1, Cdc42, and protein kinase C α (PKCα).[33] mTORC2 also phosphorylates the serine/threonine protein kinase Akt/PKB on serine residue Ser473, thus affecting metabolism and survival.[34] Phosphorylation of Akt's serine residue Ser473 by mTORC2 stimulates Akt phosphorylation on threonine residue Thr308 by PDK1 and leads to full Akt activation.[35][36] In addition, mTORC2 exhibits tyrosine protein kinase activity and phosphorylates the insulin-like growth factor 1 receptor (IGF-IR) and insulin receptor (InsR) on the tyrosine residues Tyr1131/1136 and Tyr1146/1151, respectively, leading to full activation of IGF-IR and InsR.[10]
Inhibition by rapamycin
Rapamycin inhibits mTORC1, and this appears to provide most of the beneficial effects of the drug (including life-span extension in animal studies). Rapamycin has a more complex effect on mTORC2, inhibiting it only in certain cell types under prolonged exposure. Disruption of mTORC2 produces the diabetic-like symptoms of decreased glucose tolerance and insensitivity to insulin.[37]
Gene deletion experiments
The mTORC2 signaling pathway is less defined than the mTORC1 signaling pathway. The functions of the components of the mTORC complexes have been studied using knockdowns and knockouts and were found to produce the following phenotypes:
- NIP7: Knockdown reduced mTORC2 activity that is indicated by decreased phosphorylation of mTORC2 substrates.[38]
- RICTOR: Overexpression leads to metastasis and knockdown inhibits growth factor-induced PKC-phosphorylation.[39] Constitutive deletion of Rictor in mice leads to embryonic lethality,[40] while tissue specific deletion leads to a variety of phenotypes; a common phenotype of Rictor deletion in liver, white adipose tissue, and pancreatic beta cells is systemic glucose intolerance and insulin resistance in one or more tissues.[37][41][42][43] Decreased Rictor expression in mice decreases male, but not female, lifespan.[44]
- mTOR: Inhibition of mTORC1 and mTORC2 by PP242 [2-(4-Amino-1-isopropyl-1H-pyrazolo[3,4-d]pyrimidin-3-yl)-1H-indol-5-ol] leads to autophagy or apoptosis; inhibition of mTORC2 alone by PP242 prevents phosphorylation of Ser-473 site on AKT and arrests the cells in G1 phase of the cell cycle.[45] Genetic reduction of mTOR expression in mice significantly increases lifespan.[46]
- PDK1: Knockout is lethal; hypomorphic allele results in smaller organ volume and organism size but normal AKT activation.[47]
- AKT: Knockout mice experience spontaneous apoptosis (AKT1), severe diabetes (AKT2), small brains (AKT3), and growth deficiency (AKT1/AKT2).[48] Mice heterozygous for AKT1 have increased lifespan.[49]
- TOR1, the S. cerevisiae orthologue of mTORC1, is a regulator of both carbon and nitrogen metabolism; TOR1 KO strains regulate response to nitrogen as well as carbon availability, indicating that it is a key nutritional transducer in yeast.[50][51]
Clinical significance
Aging
Decreased TOR activity has been found to increase life span in S. cerevisiae, C. elegans, and D. melanogaster.[52][53][54][55] The mTOR inhibitor rapamycin has been confirmed to increase lifespan in mice.[56][57][58][59][60]
It is hypothesized that some dietary regimes, like caloric restriction and methionine restriction, cause lifespan extension by decreasing mTOR activity.[52][53] Some studies have suggested that mTOR signaling may increase during aging, at least in specific tissues like adipose tissue, and rapamycin may act in part by blocking this increase.[61] An alternative theory is mTOR signaling is an example of antagonistic pleiotropy, and while high mTOR signaling is good during early life, it is maintained at an inappropriately high level in old age. CR and methionine restriction may act in part by limiting levels of essential amino acids including leucine and methionine, which are potent activators of mTOR.[62] The administration of leucine into the rat brain has been shown to decrease food intake and body weight via activation of the mTOR pathway.[63]
According to the free radical theory of aging,[64] reactive oxygen species cause damage of mitochondrial proteins and decrease ATP production. Subsequently, via ATP sensitive AMPK, the mTOR pathway is inhibited and ATP consuming protein synthesis is downregulated, since mTORC1 initiates a phosphorylation cascade activating the ribosome.[14] Hence, the proportion of damaged proteins is enhanced. Moreover, disruption of mTORC1 directly inhibits mitochondrial respiration.[65] These positive feedbacks on the aging process are counteracted by protective mechanisms: Decreased mTOR activity (among other factors) upregulates glycolysis[65] and removal of dysfunctional cellular components via autophagy.[64]
Cancer
Over-activation of mTOR signaling significantly contributes to the initiation and development of tumors and mTOR activity was found to be deregulated in many types of cancer including breast, prostate, lung, melanoma, bladder, brain, and renal carcinomas.[66] Reasons for constitutive activation are several. Among the most common are mutations in tumor suppressor PTEN gene. PTEN phosphatase negatively affects mTOR signalling through interfering with the effect of PI3K, an upstream effector of mTOR. Additionally, mTOR activity is deregulated in many cancers as a result of increased activity of PI3K or Akt.[67] Similarly, overexpression of downstream mTOR effectors 4E-BP1, S6K and eIF4E leads to poor cancer prognosis.[68] Also, mutations in TSC protein that inhibits the activity of mTOR may lead to a condition named tuberous sclerosis complex, which exhibits as benign lesions and increases the risk of renal cell carcinoma.[69]
Increasing mTOR activity was shown to drive cell cycle progression and increase cell proliferation mainly thanks to its effect on protein synthesis. Moreover, active mTOR supports tumor growth also indirectly by inhibiting autophagy.[70] Constitutively activated mTOR functions in supplying carcinoma cells with oxygen and nutrients by increasing the translation of HIF1A and supporting angiogenesis.[71] mTOR also aids in another metabolic adaptation of cancerous cells to support their increased growth rate - activation of glycolytic metabolism. Akt2, a substrate of mTOR, specifically of mTORC2, upregulates expression of the glycolytic enzyme PKM2 thus contributing to the Warburg effect.[72]
Central nervous system disorders
Autism
MTOR is implicated in the failure of a 'pruning' mechanism of the excitatory synapses in autism spectrum disorders.[73]
Alzheimer’s disease
mTOR signaling intersects with Alzheimer’s disease (AD) pathology in several aspects, suggesting its potential role as a contributor to disease progression. In general, findings demonstrate mTOR signaling hyperactivity in AD brains. For example, postmortem studies of human AD brain reveal dysregulation in PTEN, Akt, S6K, and mTOR.[74][75][76] mTOR signaling appears to be closely related to the presence of soluble amyloid beta (Aβ) and tau proteins, which aggregate and form two hallmarks of the disease, Aβ plaques and neurofibrillary tangles, respectively.[77] In vitro studies have shown Aβ to be an activator of the PI3K/AKT pathway, which in turn activates mTOR.[78] In addition, applying Aβ to N2K cells increases the expression of p70S6K, a downstream target of mTOR known to have higher expression in neurons that eventually develop neurofibrillary tangles.[79][80] Chinese hamster ovary cells transfected with the 7PA2 familial AD mutation also exhibit increased mTOR activity compared to controls, and the hyperactivity is blocked using a gamma-secretase inhibitor.[81][82] These in vitro studies suggest that increasing Aβ concentrations increases mTOR signaling; however, significantly large, cytotoxic Aβ concentrations are thought to decrease mTOR signaling.[83]
Consistent with data observed in vitro, mTOR activity and activated p70S6K have been shown to be significantly increased in the cortex and hippocampus of animal models of AD compared to controls.[82][84] Pharmacologic or genetic removal of the Aβ in animal models of AD eliminates the disruption in normal mTOR activity, pointing to the direct involvement of Aβ in mTOR signaling.[84] In addition, by injecting Aβ oligomers into the hippocampi of normal mice, mTOR hyperactivity is observed.[84] Cognitive impairments characteristic of AD appear to be mediated by the phosphorylation of PRAS-40, which detaches from and allows for the mTOR hyperactivity when it is phosphorylated; inhibiting PRAS-40 phosphorylation prevents Aβ-induced mTOR hyperactivity.[84][85][86] Given these findings, the mTOR signaling pathway appears to be one mechanism of Aβ-induced toxicity in AD.
The hyperphosphorylation of tau proteins into neurofibrillary tangles is one hallmark of AD. p70S6K activation has been shown to promote tangle formation as well as mTOR hyperactivity through increased phosphorylation and reduced dephosphorylation.[79][87][88][89] It has also been proposed that mTOR contributes to tau pathology by increasing the translation of tau and other proteins.[90]
Synaptic plasticity is a key contributor to learning and memory, two processes that are severely impaired in AD patients. Translational control, or the maintenance of protein homeostasis, has been shown to be essential for neural plasticity and is regulated by mTOR.[82][91][92][93][94] Both protein over- and under-production via mTOR activity seem to contribute to impaired learning and memory. Furthermore, given that deficits resulting from mTOR overactivity can be alleviated through treatment with rapamycin, it is possible that mTOR plays an important role in affecting cognitive functioning through synaptic plasticity.[78][95] Further evidence for mTOR activity in neurodegeneration comes from recent findings demonstrating that eIF2α-P, an upstream target of the mTOR pathway, mediates cell death in prion diseases through sustained translational inhibition.[96]
Some evidence points to mTOR’s role in reduced Aβ clearance as well. mTOR is a negative regulator of autophagy;[97] therefore, hyperactivity in mTOR signaling should reduce Aβ clearance in the AD brain. Disruptions in autophagy may be a potential source of pathogenesis in protein misfolding diseases, including AD.[98][99][100][101][102][103] Studies using mouse models of Huntington’s disease demonstrate that treatment with rapamycin facilitates the clearance of huntingtin aggregates.[104][105] Perhaps the same treatment may be useful in clearing Aβ deposits as well.
Protein synthesis and cell growth
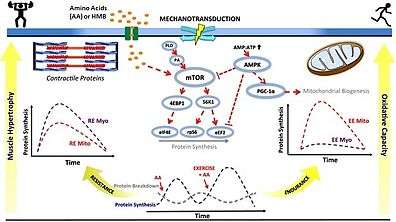
mTORC1 activation is required for myofibrillar muscle protein synthesis and skeletal muscle hypertrophy in humans in response to both physical exercise and ingestion of certain amino acids or amino acid derivatives.[106][107] Persistent inactivation of mTORC1 signaling in skeletal muscle facilitates the loss of muscle mass and strength during muscle wasting in old age, cancer cachexia, and muscle atrophy from physical inactivity.[106][107][108] mTORC2 activation appears to mediate neurite outgrowth in differentiated mouse neuro2a cells.[109] Intermittent mTOR activation in prefrontal neurons by β-hydroxy β-methylbutyrate inhibits age-related cognitive decline associated with dendritic pruning in animals, which is a phenomenon also observed in humans.[110]
• PA: phosphatidic acid
• mTOR: mechanistic target of rapamycin
• AMP: adenosine monophosphate
• ATP: adenosine triphosphate
• AMPK: AMP-activated protein kinase
• PGC‐1α: peroxisome proliferator-activated receptor gamma coactivator-1α
• S6K1: p70S6 kinase
• 4EBP1: eukaryotic translation initiation factor 4E-binding protein 1
• eIF4E: eukaryotic translation initiation factor 4E
• RPS6: ribosomal protein S6
• eEF2: eukaryotic elongation factor 2
• RE: resistance exercise; EE: endurance exercise
• Myo: myofibrillar; Mito: mitochondrial
• AA: amino acids
• HMB: β-hydroxy β-methylbutyric acid
• ↑ represents activation
• Τ represents inhibition
Scleroderma
Scleroderma, also known as systemic sclerosis, is a chronic systemic autoimmune disease characterised by hardening (sclero) of the skin (derma) that affects internal organs in its more severe forms.[112][113] mTOR plays a role in fibrotic diseases and autoimmunity, and blockade of the mTORC pathway is under investigation as a treatment for scleroderma.[7]
mTOR inhibitors as therapies
mTOR inhibitors, e.g. rapamycin, are already used to prevent transplant rejection. Rapamycin is also related to the therapy of glycogen storage disease (GSD). Some articles reported that rapamycin can inhibit mTORC1 so that the phosphorylation of GS(glycogen synthase) can be increased in skeletal muscle. This discovery represents a potential novel therapeutic approach for glycogen storage diseases that involve glycogen accumulation in muscle. Various natural compounds, including epigallocatechin gallate (EGCG), caffeine, curcumin, and resveratrol, have also been reported to inhibit mTOR when applied to isolated cells in culture;[22][114] however, there is as yet no evidence that these substances inhibit mTOR when taken as dietary supplements.
Some mTOR inhibitors (e.g. temsirolimus, everolimus) are beginning to be used in the treatment of cancer.[69][115] mTOR inhibitors may also be useful for treating several age-associated diseases[116] including neurodegenerative diseases such as Alzheimer's disease and Parkinson's disease.[117] Ridaforolimus is another mTOR inhibitor, currently in clinical development.
Interactions
Mammalian target of rapamycin has been shown to interact with:[118]
- ABL1,[119]
- AKT1,[35][120][121]
- IGF-IR,[10]
- InsR,[10]
- CLIP1,[122]
- EIF3F[123]
- EIF4EBP1,[28][124][125][126][127][128][129][130]
- FKBP1A,[11][33][131][132][133][134]
- GPHN,[135]
- KIAA1303,[11][28][32][33][65][124][125][126][136][137][138][139][140][141][142][143][144][145][146][147]
- PRKCD,[148]
- RHEB,[127][149][150][151]
- RICTOR,[11][32][33][138][144][146][147]
- RPS6KB1,[28][125][127][128][129][143][146][152][153][154][155][156][157][157][158][159]
- STAT1,[160]
- STAT3,[161][162]
- Two-pore channels: TPCN1; TPCN2,[163] and
- UBQLN1.[164]
References
- ↑ "Drugs that physically interact with Serine/threonine-protein kinase mTOR view/edit references on wikidata".
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Lamming DW, Ye L, Sabatini DM, Baur JA (2013). "Rapalogs and mTOR inhibitors as anti-aging therapeutics". Journal of Clinical Investigation. 123 (3): 980–989. doi:10.1172/JCI64099. PMC 3582126
 . PMID 23454761.
. PMID 23454761. - 1 2 Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL (June 1994). "A mammalian protein targeted by G1-arresting rapamycin-receptor complex". Nature. 369 (6483): 756–8. doi:10.1038/369756a0. PMID 8008069.
- ↑ Moore PA, Rosen CA, Carter KC (April 1996). "Assignment of the human FKBP12-rapamycin-associated protein (FRAP) gene to chromosome 1p36 by fluorescence in situ hybridization". Genomics. 33 (2): 331–2. doi:10.1006/geno.1996.0206. PMID 8660990.
- 1 2 Mitra A, Luna JI, Marusina AI, Merleev A, Kundu-Raychaudhuri S, Fiorentino D, Raychaudhuri SP, Maverakis E (November 2015). "Dual mTOR Inhibition Is Required to Prevent TGF-β-Mediated Fibrosis: Implications for Scleroderma". The Journal of Investigative Dermatology. 135 (11): 2873–6. doi:10.1038/jid.2015.252. PMC 4640976
 . PMID 26134944.
. PMID 26134944. - 1 2 3 4 5 6 7 Lipton JO, Sahin M (October 2014). "The neurology of mTOR". Neuron. 84 (2): 275–291. doi:10.1016/j.neuron.2014.09.034. PMC 4223653
 . PMID 25374355.
. PMID 25374355. The mTOR signaling pathway acts as a molecular systems integrator to support organismal and cellular interactions with the environment. The mTOR pathway regulates homeostasis by directly influencing protein synthesis, transcription, autophagy, metabolism, and organelle biogenesis and maintenance. It is not surprising then that mTOR signaling is implicated in the entire hierarchy of brain function including the proliferation of neural stem cells, the assembly and maintenance of circuits, experience-dependent plasticity and regulation of complex behaviors like feeding, sleep and circadian rhythms. ...
mTOR function is mediated through two large biochemical complexes defined by their respective protein composition and have been extensively reviewed elsewhere(Dibble and Manning, 2013; Laplante and Sabatini, 2012)(Figure 1B). In brief, common to both mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) are: mTOR itself, mammalian lethal with sec13 protein 8 (mLST8; also known as GβL), and the inhibitory DEP domain containing mTOR-interacting protein (DEPTOR). Specific to mTORC1 is the regulator-associated protein of the mammalian target of rapamycin (Raptor) and proline-rich Akt substrate of 40 kDa (PRAS40)(Kim et al., 2002; Laplante and Sabatini, 2012). Raptor is essential to mTORC1 activity. The mTORC2 complex includes the rapamycin insensitive companion of mTOR (Rictor), mammalian stress activated MAP kinase-interacting protein 1 (mSIN1), and proteins observed with rictor 1 and 2 (PROTOR 1 and 2)(Jacinto et al., 2006; Jacinto et al., 2004; Pearce et al., 2007; Sarbassov et al., 2004)(Figure 1B). Rictor and mSIN1 are both critical to mTORC2 function.
Figure 1: Domain structure of the mTOR kinase and components of mTORC1 and mTORC2
Figure 2: The mTOR Signaling Pathway - 1 2 3 4 Hay N, Sonenberg N (August 2004). "Upstream and downstream of mTOR". Genes & Development. 18 (16): 1926–45. doi:10.1101/gad.1212704. PMID 15314020.
- 1 2 3 4 Yin Y, Hua H, Li M, Liu S, Kong Q, Shao T, Wang J, Luo Y, Wang Q, Luo T, Jiang Y (January 2016). "mTORC2 promotes type I insulin-like growth factor receptor and insulin receptor activation through the tyrosine kinase activity of mTOR". Cell Research. 26 (1): 46–65. doi:10.1038/cr.2015.133. PMID 26584640.
- 1 2 3 4 Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN (November 2004). "Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive". Nature Cell Biology. 6 (11): 1122–8. doi:10.1038/ncb1183. PMID 15467718.
- ↑ Vézina C, Kudelski A, Sehgal SN (October 1975). "Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle". The Journal of Antibiotics. 28 (10): 721–6. doi:10.7164/antibiotics.28.721. PMID 1102508.
- ↑ Dobashi Y, Watanabe Y, Miwa C, Suzuki S, Koyama S (June 2011). "Mammalian target of rapamycin: a central node of complex signaling cascades". International Journal of Clinical and Experimental Pathology. 4 (5): 476–95. PMC 3127069
 . PMID 21738819.
. PMID 21738819. - 1 2 Magnuson B, Ekim B, Fingar DC (January 2012). "Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks". The Biochemical Journal. 441 (1): 1–21. doi:10.1042/BJ20110892. PMID 22168436.
- ↑ Abraham RT, Wiederrecht GJ (1996). "Immunopharmacology of rapamycin". Annual Review of Immunology. 14: 483–510. doi:10.1146/annurev.immunol.14.1.483. PMID 8717522.
- ↑ Heitman J, Movva NR, Hall MN (August 1991). "Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast". Science. 253 (5022): 905–9. doi:10.1126/science.1715094. PMID 1715094.
- ↑ Heitman J (2015-11-05). "On the Discovery of TOR As the Target of Rapamycin". PLoS Pathogens. 11 (11): e1005245. doi:10.1371/journal.ppat.1005245. PMC 4634758
 . PMID 26540102.
. PMID 26540102. - ↑ Foster KG, Fingar DC (May 2010). "Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony". The Journal of Biological Chemistry. 285 (19): 14071–7. doi:10.1074/jbc.R109.094003. PMC 2863215
 . PMID 20231296.
. PMID 20231296. - ↑ Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH (July 1994). "RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs". Cell. 78 (1): 35–43. doi:10.1016/0092-8674(94)90570-3. PMID 7518356.
- ↑ Pópulo H, Lopes JM, Soares P (10 February 2012). "The mTOR signalling pathway in human cancer". International Journal of Molecular Sciences. 13 (2): 1886–918. doi:10.3390/ijms13021886. PMC 3291999
 . PMID 22408430.
. PMID 22408430. - ↑ Tokunaga C, Yoshino K, Yonezawa K (January 2004). "mTOR integrates amino acid- and energy-sensing pathways". Biochemical and Biophysical Research Communications. 313 (2): 443–6. doi:10.1016/j.bbrc.2003.07.019. PMID 14684182.
- 1 2 Beevers CS, Li F, Liu L, Huang S (August 2006). "Curcumin inhibits the mammalian target of rapamycin-mediated signaling pathways in cancer cells". International Journal of Cancer. 119 (4): 757–64. doi:10.1002/ijc.21932. PMID 16550606.
- ↑ Kennedy BK, Lamming DW (June 2016). "The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging". Cell Metabolism. 23 (6): 990–1003. doi:10.1016/j.cmet.2016.05.009. PMC 4910876
 . PMID 27304501.
. PMID 27304501. - ↑ Huang S, Houghton PJ (December 2001). "Mechanisms of resistance to rapamycins". Drug Resistance Updates. 4 (6): 378–91. doi:10.1054/drup.2002.0227. PMID 12030785.
- 1 2 Huang S, Bjornsti MA, Houghton PJ (2003). "Rapamycins: mechanism of action and cellular resistance". Cancer Biology & Therapy. 2 (3): 222–32. doi:10.4161/cbt.2.3.360. PMID 12878853.
- ↑ Wullschleger S, Loewith R, Hall MN (February 2006). "TOR signaling in growth and metabolism". Cell. 124 (3): 471–84. doi:10.1016/j.cell.2006.01.016. PMID 16469695.
- ↑ Betz C, Hall MN (November 2013). "Where is mTOR and what is it doing there?". The Journal of Cell Biology. 203 (4): 563–74. doi:10.1083/jcb.201306041. PMID 24385483.
- 1 2 3 4 5 6 Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (July 2002). "mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery". Cell. 110 (2): 163–75. doi:10.1016/S0092-8674(02)00808-5. PMID 12150925.
- ↑ Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM (April 2003). "GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR". Molecular Cell. 11 (4): 895–904. doi:10.1016/S1097-2765(03)00114-X. PMID 12718876.
- ↑ Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J (November 2001). "Phosphatidic acid-mediated mitogenic activation of mTOR signaling". Science. 294 (5548): 1942–5. doi:10.1126/science.1066015. PMID 11729323.
- ↑ Bond P (March 2016). "Regulation of mTORC1 by growth factors, energy status, amino acids and mechanical stimuli at a glance". J. Int. Soc. Sports Nutr. 13: 8. doi:10.1186/s12970-016-0118-y. PMC 4774173
 . PMID 26937223.
. PMID 26937223. - 1 2 3 Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM (September 2006). "mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s". Current Biology. 16 (18): 1865–70. doi:10.1016/j.cub.2006.08.001. PMID 16919458.
- 1 2 3 4 5 Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM (July 2004). "Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton". Current Biology. 14 (14): 1296–302. doi:10.1016/j.cub.2004.06.054. PMID 15268862.
- ↑ Betz C, Stracka D, Prescianotto-Baschong C, Frieden M, Demaurex N, Hall MN (July 2013). "Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology". Proceedings of the National Academy of Sciences of the United States of America. 110 (31): 12526–34. doi:10.1073/pnas.1302455110. PMC 3732980
 . PMID 23852728.
. PMID 23852728. - 1 2 Sarbassov DD, Guertin DA, Ali SM, Sabatini DM (February 2005). "Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex". Science. 307 (5712): 1098–101. doi:10.1126/science.1106148. PMID 15718470.
- ↑ Stephens L, Anderson K, Stokoe D, Erdjument-Bromage H, Painter GF, Holmes AB, Gaffney PR, Reese CB, McCormick F, Tempst P, Coadwell J, Hawkins PT (January 1998). "Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B". Science. 279 (5351): 710–4. doi:10.1126/science.279.5351.710. PMID 9445477.
- 1 2 Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, Guertin DA, Sabatini DM, Baur JA (March 2012). "Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity". Science. 335 (6076): 1638–43. doi:10.1126/science.1215135. PMC 3324089
 . PMID 22461615.
. PMID 22461615. - ↑ Zinzalla V, Stracka D, Oppliger W, Hall MN (March 2011). "Activation of mTORC2 by association with the ribosome". Cell. 144 (5): 757–68. doi:10.1016/j.cell.2011.02.014. PMID 21376236.
- ↑ Zhang F, Zhang X, Li M, Chen P, Zhang B, Guo H, Cao W, Wei X, Cao X, Hao X, Zhang N (November 2010). "mTOR complex component Rictor interacts with PKCzeta and regulates cancer cell metastasis". Cancer Research. 70 (22): 9360–70. doi:10.1158/0008-5472.CAN-10-0207. PMID 20978191.
- ↑ Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM (December 2006). "Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1". Developmental Cell. 11 (6): 859–71. doi:10.1016/j.devcel.2006.10.007. PMID 17141160.
- ↑ Gu Y, Lindner J, Kumar A, Yuan W, Magnuson MA (March 2011). "Rictor/mTORC2 is essential for maintaining a balance between beta-cell proliferation and cell size". Diabetes. 60 (3): 827–37. doi:10.2337/db10-1194. PMC 3046843
 . PMID 21266327.
. PMID 21266327. - ↑ Lamming DW, Demirkan G, Boylan JM, Mihaylova MM, Peng T, Ferreira J, Neretti N, Salomon A, Sabatini DM, Gruppuso PA (January 2014). "Hepatic signaling by the mechanistic target of rapamycin complex 2 (mTORC2)". FASEB Journal. 28 (1): 300–15. doi:10.1096/fj.13-237743. PMC 3868844
 . PMID 24072782.
. PMID 24072782. - ↑ Kumar A, Lawrence JC, Jung DY, Ko HJ, Keller SR, Kim JK, Magnuson MA, Harris TE (June 2010). "Fat cell-specific ablation of rictor in mice impairs insulin-regulated fat cell and whole-body glucose and lipid metabolism". Diabetes. 59 (6): 1397–406. doi:10.2337/db09-1061. PMC 2874700
 . PMID 20332342.
. PMID 20332342. - ↑ Lamming DW, Mihaylova MM, Katajisto P, Baar EL, Yilmaz OH, Hutchins A, Gultekin Y, Gaither R, Sabatini DM (October 2014). "Depletion of Rictor, an essential protein component of mTORC2, decreases male lifespan". Aging Cell. 13 (5): 911–7. doi:10.1111/acel.12256. PMC 4172536
 . PMID 25059582.
. PMID 25059582. - ↑ Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM (February 2009). Hunter T, ed. "Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2". PLoS Biology. 7 (2): e38. doi:10.1371/journal.pbio.1000038. PMC 2637922
 . PMID 19209957.
. PMID 19209957. - ↑ Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, Fergusson MM, Rovira II, Allen M, Springer DA, Lago CU, Zhang S, DuBois W, Ward T, deCabo R, Gavrilova O, Mock B, Finkel T (September 2013). "Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression". Cell Reports. 4 (5): 913–20. doi:10.1016/j.celrep.2013.07.030. PMC 3784301
 . PMID 23994476.
. PMID 23994476. - ↑ Lawlor MA, Mora A, Ashby PR, Williams MR, Murray-Tait V, Malone L, Prescott AR, Lucocq JM, Alessi DR (July 2002). "Essential role of PDK1 in regulating cell size and development in mice". The EMBO Journal. 21 (14): 3728–38. doi:10.1093/emboj/cdf387. PMC 126129
 . PMID 12110585.
. PMID 12110585. - ↑ Yang ZZ, Tschopp O, Baudry A, Dümmler B, Hynx D, Hemmings BA (April 2004). "Physiological functions of protein kinase B/Akt". Biochemical Society Transactions. 32 (Pt 2): 350–4. doi:10.1042/BST0320350. PMID 15046607.
- ↑ Nojima A, Yamashita M, Yoshida Y, Shimizu I, Ichimiya H, Kamimura N, Kobayashi Y, Ohta S, Ishii N, Minamino T (2013-01-01). "Haploinsufficiency of akt1 prolongs the lifespan of mice". PloS One. 8 (7): e69178. doi:10.1371/journal.pone.0069178. PMC 3728301
 . PMID 23935948.
. PMID 23935948. - ↑ Crespo JL, Hall MN (December 2002). "Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae". Microbiology and Molecular Biology Reviews. 66 (4): 579–91, table of contents. doi:10.1128/mmbr.66.4.579-591.2002. PMC 134654
 . PMID 12456783.
. PMID 12456783. - ↑ Peter GJ, Düring L, Ahmed A (March 2006). "Carbon catabolite repression regulates amino acid permeases in Saccharomyces cerevisiae via the TOR signaling pathway". The Journal of Biological Chemistry. 281 (9): 5546–52. doi:10.1074/jbc.M513842200. PMID 16407266.
- 1 2 Powers RW, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S (January 2006). "Extension of chronological life span in yeast by decreased TOR pathway signaling". Genes & Development. 20 (2): 174–84. doi:10.1101/gad.1381406. PMC 1356109
 . PMID 16418483.
. PMID 16418483. - 1 2 Kaeberlein M, Powers RW, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK (November 2005). "Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients". Science. 310 (5751): 1193–6. doi:10.1126/science.1115535. PMID 16293764.
- ↑ Jia K, Chen D, Riddle DL (August 2004). "The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span". Development. 131 (16): 3897–906. doi:10.1242/dev.01255. PMID 15253933.
- ↑ Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S (May 2004). "Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway". Current Biology. 14 (10): 885–90. doi:10.1016/j.cub.2004.03.059. PMC 2754830
 . PMID 15186745.
. PMID 15186745. - ↑ Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA (July 2009). "Rapamycin fed late in life extends lifespan in genetically heterogeneous mice". Nature. 460 (7253): 392–5. doi:10.1038/nature08221. PMC 2786175
 . PMID 19587680.
. PMID 19587680. - ↑ Miller RA, Harrison DE, Astle CM, Fernandez E, Flurkey K, Han M, Javors MA, Li X, Nadon NL, Nelson JF, Pletcher S, Salmon AB, Sharp ZD, Van Roekel S, Winkleman L, Strong R (June 2014). "Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction". Aging Cell. 13 (3): 468–77. doi:10.1111/acel.12194. PMC 4032600
 . PMID 24341993.
. PMID 24341993. - ↑ Fok WC, Chen Y, Bokov A, Zhang Y, Salmon AB, Diaz V, Javors M, Wood WH, Zhang Y, Becker KG, Pérez VI, Richardson A (2014-01-01). "Mice fed rapamycin have an increase in lifespan associated with major changes in the liver transcriptome". PloS One. 9 (1): e83988. doi:10.1371/journal.pone.0083988. PMC 3883653
 . PMID 24409289.
. PMID 24409289. - ↑ Arriola Apelo SI, Pumper CP, Baar EL, Cummings NE, Lamming DW (July 2016). "Intermittent Administration of Rapamycin Extends the Life Span of Female C57BL/6J Mice". The Journals of Gerontology. Series A, Biological Sciences and Medical Sciences. 71 (7): 876–81. doi:10.1093/gerona/glw064. PMID 27091134.
- ↑ Popovich IG, Anisimov VN, Zabezhinski MA, Semenchenko AV, Tyndyk ML, Yurova MN, Blagosklonny MV (May 2014). "Lifespan extension and cancer prevention in HER-2/neu transgenic mice treated with low intermittent doses of rapamycin". Cancer Biology & Therapy. 15 (5): 586–92. doi:10.4161/cbt.28164. PMC 4026081
 . PMID 24556924.
. PMID 24556924. - ↑ Baar EL, Carbajal KA, Ong IM, Lamming DW (February 2016). "Sex- and tissue-specific changes in mTOR signaling with age in C57BL/6J mice". Aging Cell. 15 (1): 155–66. doi:10.1111/acel.12425. PMC 4717274
 . PMID 26695882.
. PMID 26695882. - ↑ Caron A, Richard D, Laplante M (Jul 2015). "The Roles of mTOR Complexes in Lipid Metabolism". Annual Review of Nutrition. 35: 321–48. doi:10.1146/annurev-nutr-071714-034355. PMID 26185979.
- ↑ Cota D, Proulx K, Smith KA, Kozma SC, Thomas G, Woods SC, Seeley RJ (May 2006). "Hypothalamic mTOR signaling regulates food intake". Science. 312 (5775): 927–30. doi:10.1126/science.1124147. PMID 16690869.
- 1 2 Kriete A, Bosl WJ, Booker G (June 2010). "Rule-based cell systems model of aging using feedback loop motifs mediated by stress responses". PLoS Computational Biology. 6 (6): e1000820. doi:10.1371/journal.pcbi.1000820. PMC 2887462
 . PMID 20585546.
. PMID 20585546. - 1 2 3 Schieke SM, Phillips D, McCoy JP, Aponte AM, Shen RF, Balaban RS, Finkel T (September 2006). "The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity". The Journal of Biological Chemistry. 281 (37): 27643–52. doi:10.1074/jbc.M603536200. PMID 16847060.
- ↑ Xu K, Liu P, Wei W (December 2014). "mTOR signaling in tumorigenesis". Biochimica et Biophysica Acta. 1846 (2): 638–54. doi:10.1016/j.bbcan.2014.10.007. PMID 25450580.
- ↑ Guertin DA, Sabatini DM (August 2005). "An expanding role for mTOR in cancer". Trends in Molecular Medicine. 11 (8): 353–61. doi:10.1016/j.molmed.2005.06.007. PMID 16002336.
- ↑ Pópulo H, Lopes JM, Soares P (2012). "The mTOR signalling pathway in human cancer". International Journal of Molecular Sciences. 13 (2): 1886–918. doi:10.3390/ijms13021886. PMC 3291999
 . PMID 22408430.
. PMID 22408430. - 1 2 Easton JB, Houghton PJ (October 2006). "mTOR and cancer therapy". Oncogene. 25 (48): 6436–46. doi:10.1038/sj.onc.1209886. PMID 17041628.
- ↑ Zoncu R, Efeyan A, Sabatini DM (January 2011). "mTOR: from growth signal integration to cancer, diabetes and ageing". Nature Reviews Molecular Cell Biology. 12 (1): 21–35. doi:10.1038/nrm3025. PMC 3390257
 . PMID 21157483.
. PMID 21157483. - ↑ Thomas GV, Tran C, Mellinghoff IK, Welsbie DS, Chan E, Fueger B, Czernin J, Sawyers CL (January 2006). "Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer". Nature Medicine. 12 (1): 122–7. doi:10.1038/nm1337. PMID 16341243.
- ↑ Nemazanyy I, Espeillac C, Pende M, Panasyuk G (August 2013). "Role of PI3K, mTOR and Akt2 signalling in hepatic tumorigenesis via the control of PKM2 expression". Biochemical Society Transactions. 41 (4): 917–22. doi:10.1042/BST20130034. PMID 23863156.
- ↑ Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, Yue Z, Arancio O, Peterson BS, Champagne F, Dwork AJ, Goldman J, Sulzer D (September 2014). "Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits". Neuron. 83 (5): 1131–43. doi:10.1016/j.neuron.2014.07.040. PMID 25155956.
- ↑ Rosner M, Hanneder M, Siegel N, Valli A, Fuchs C, Hengstschläger M (June 2008). "The mTOR pathway and its role in human genetic diseases". Mutation Research. 659 (3): 284–92. doi:10.1016/j.mrrev.2008.06.001. PMID 18598780.
- ↑ Li X, Alafuzoff I, Soininen H, Winblad B, Pei JJ (August 2005). "Levels of mTOR and its downstream targets 4E-BP1, eEF2, and eEF2 kinase in relationships with tau in Alzheimer's disease brain". The FEBS Journal. 272 (16): 4211–20. doi:10.1111/j.1742-4658.2005.04833.x. PMID 16098202.
- ↑ Chano T, Okabe H, Hulette CM (September 2007). "RB1CC1 insufficiency causes neuronal atrophy through mTOR signaling alteration and involved in the pathology of Alzheimer's diseases". Brain Research. 1168 (1168): 97–105. doi:10.1016/j.brainres.2007.06.075. PMID 17706618.
- ↑ Selkoe DJ (September 2008). "Soluble oligomers of the amyloid beta-protein impair synaptic plasticity and behavior". Behavioural Brain Research. 192 (1): 106–13. doi:10.1016/j.bbr.2008.02.016. PMC 2601528
 . PMID 18359102.
. PMID 18359102. - 1 2 Oddo S (January 2012). "The role of mTOR signaling in Alzheimer disease". Frontiers in Bioscience. 4 (1): 941–52. doi:10.2741/s310. PMID 22202101.
- 1 2 An WL, Cowburn RF, Li L, Braak H, Alafuzoff I, Iqbal K, Iqbal IG, Winblad B, Pei JJ (August 2003). "Up-regulation of phosphorylated/activated p70 S6 kinase and its relationship to neurofibrillary pathology in Alzheimer's disease". The American Journal of Pathology. 163 (2): 591–607. doi:10.1016/S0002-9440(10)63687-5. PMC 1868198
 . PMID 12875979.
. PMID 12875979. - ↑ Zhang F, Beharry ZM, Harris TE, Lilly MB, Smith CD, Mahajan S, Kraft AS (May 2009). "PIM1 protein kinase regulates PRAS40 phosphorylation and mTOR activity in FDCP1 cells". Cancer Biology & Therapy. 8 (9): 846–53. doi:10.4161/cbt.8.9.8210. PMID 19276681.
- ↑ Koo EH, Squazzo SL (July 1994). "Evidence that production and release of amyloid beta-protein involves the endocytic pathway". The Journal of Biological Chemistry. 269 (26): 17386–9. PMID 8021238.
- 1 2 3 Caccamo A, Majumder S, Richardson A, Strong R, Oddo S (April 2010). "Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-beta, and Tau: effects on cognitive impairments". The Journal of Biological Chemistry. 285 (17): 13107–20. doi:10.1074/jbc.M110.100420. PMC 2857107
 . PMID 20178983.
. PMID 20178983. - ↑ Lafay-Chebassier C, Paccalin M, Page G, Barc-Pain S, Perault-Pochat MC, Gil R, Pradier L, Hugon J (July 2005). "mTOR/p70S6k signalling alteration by Abeta exposure as well as in APP-PS1 transgenic models and in patients with Alzheimer's disease". Journal of Neurochemistry. 94 (1): 215–25. doi:10.1111/j.1471-4159.2005.03187.x. PMID 15953364.
- 1 2 3 4 Caccamo A, Maldonado MA, Majumder S, Medina DX, Holbein W, Magrí A, Oddo S (March 2011). "Naturally secreted amyloid-beta increases mammalian target of rapamycin (mTOR) activity via a PRAS40-mediated mechanism". The Journal of Biological Chemistry. 286 (11): 8924–32. doi:10.1074/jbc.M110.180638. PMC 3058958
 . PMID 21266573.
. PMID 21266573. - ↑ Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM (March 2007). "PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase". Molecular Cell. 25 (6): 903–15. doi:10.1016/j.molcel.2007.03.003. PMID 17386266.
- ↑ Wang L, Harris TE, Roth RA, Lawrence JC (July 2007). "PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding". The Journal of Biological Chemistry. 282 (27): 20036–44. doi:10.1074/jbc.M702376200. PMID 17510057.
- ↑ Pei JJ, Hugon J (December 2008). "mTOR-dependent signalling in Alzheimer's disease". Journal of Cellular and Molecular Medicine. 12 (6B): 2525–32. doi:10.1111/j.1582-4934.2008.00509.x. PMID 19210753.
- ↑ Meske V, Albert F, Ohm TG (January 2008). "Coupling of mammalian target of rapamycin with phosphoinositide 3-kinase signaling pathway regulates protein phosphatase 2A- and glycogen synthase kinase-3 -dependent phosphorylation of Tau". The Journal of Biological Chemistry. 283 (1): 100–9. doi:10.1074/jbc.M704292200. PMID 17971449.
- ↑ Janssens V, Goris J (February 2001). "Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling". The Biochemical Journal. 353 (Pt 3): 417–39. doi:10.1042/0264-6021:3530417. PMC 1221586
 . PMID 11171037.
. PMID 11171037. - ↑ Morita T, Sobue K (October 2009). "Specification of neuronal polarity regulated by local translation of CRMP2 and Tau via the mTOR-p70S6K pathway". The Journal of Biological Chemistry. 284 (40): 27734–45. doi:10.1074/jbc.M109.008177. PMC 2785701
 . PMID 19648118.
. PMID 19648118. - ↑ Puighermanal E, Marsicano G, Busquets-Garcia A, Lutz B, Maldonado R, Ozaita A (September 2009). "Cannabinoid modulation of hippocampal long-term memory is mediated by mTOR signaling". Nature Neuroscience. 12 (9): 1152–8. doi:10.1038/nn.2369. PMID 19648913.
- ↑ Tischmeyer W, Schicknick H, Kraus M, Seidenbecher CI, Staak S, Scheich H, Gundelfinger ED (August 2003). "Rapamycin-sensitive signalling in long-term consolidation of auditory cortex-dependent memory". The European Journal of Neuroscience. 18 (4): 942–50. doi:10.1046/j.1460-9568.2003.02820.x. PMID 12925020.
- ↑ Hoeffer CA, Klann E (February 2010). "mTOR signaling: at the crossroads of plasticity, memory and disease". Trends in Neurosciences. 33 (2): 67–75. doi:10.1016/j.tins.2009.11.003. PMC 2821969
 . PMID 19963289.
. PMID 19963289. - ↑ Kelleher RJ, Govindarajan A, Jung HY, Kang H, Tonegawa S (February 2004). "Translational control by MAPK signaling in long-term synaptic plasticity and memory". Cell. 116 (3): 467–79. doi:10.1016/S0092-8674(04)00115-1. PMID 15016380.
- ↑ Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, Kwiatkowski DJ, Ramesh V, Silva AJ (August 2008). "Reversal of learning deficits in a Tsc2+/- mouse model of tuberous sclerosis". Nature Medicine. 14 (8): 843–8. doi:10.1038/nm1788. PMC 2664098
 . PMID 18568033.
. PMID 18568033. - ↑ Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, Halliday M, Morgan J, Dinsdale D, Ortori CA, Barrett DA, Tsaytler P, Bertolotti A, Willis AE, Bushell M, Mallucci GR (May 2012). "Sustained translational repression by eIF2α-P mediates prion neurodegeneration". Nature. 485 (7399): 507–11. doi:10.1038/nature11058. PMC 3378208
 . PMID 22622579.
. PMID 22622579. - ↑ Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL (October 2008). "The role of TOR in autophagy regulation from yeast to plants and mammals". Autophagy. 4 (7): 851–65. doi:10.4161/auto.6555. PMID 18670193.
- ↑ McCray BA, Taylor JP (December 2008). "The role of autophagy in age-related neurodegeneration". Neuro-Signals. 16 (1): 75–84. doi:10.1159/000109761. PMID 18097162.
- ↑ Nedelsky NB, Todd PK, Taylor JP (December 2008). "Autophagy and the ubiquitin-proteasome system: collaborators in neuroprotection". Biochimica et Biophysica Acta. 1782 (12): 691–9. doi:10.1016/j.bbadis.2008.10.002. PMC 2621359
 . PMID 18930136.
. PMID 18930136. - ↑ Rubinsztein DC (October 2006). "The roles of intracellular protein-degradation pathways in neurodegeneration". Nature. 443 (7113): 780–6. doi:10.1038/nature05291. PMID 17051204.
- ↑ Oddo S (April 2008). "The ubiquitin-proteasome system in Alzheimer's disease". Journal of Cellular and Molecular Medicine. 12 (2): 363–73. doi:10.1111/j.1582-4934.2008.00276.x. PMID 18266959.
- ↑ Li X, Li H, Li XJ (November 2008). "Intracellular degradation of misfolded proteins in polyglutamine neurodegenerative diseases". Brain Research Reviews. 59 (1): 245–52. doi:10.1016/j.brainresrev.2008.08.003. PMC 2577582
 . PMID 18773920.
. PMID 18773920. - ↑ Caccamo A, Majumder S, Deng JJ, Bai Y, Thornton FB, Oddo S (October 2009). "Rapamycin rescues TDP-43 mislocalization and the associated low molecular mass neurofilament instability". The Journal of Biological Chemistry. 284 (40): 27416–24. doi:10.1074/jbc.M109.031278. PMC 2785671
 . PMID 19651785.
. PMID 19651785. - ↑ Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, Rubinsztein DC (June 2004). "Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease". Nature Genetics. 36 (6): 585–95. doi:10.1038/ng1362. PMID 15146184.
- ↑ Rami A (October 2009). "Review: autophagy in neurodegeneration: firefighter and/or incendiarist?". Neuropathology and Applied Neurobiology. 35 (5): 449–61. doi:10.1111/j.1365-2990.2009.01034.x. PMID 19555462.
- 1 2 3 4 Brook MS, Wilkinson DJ, Phillips BE, Perez-Schindler J, Philp A, Smith K, Atherton PJ (January 2016). "Skeletal muscle homeostasis and plasticity in youth and ageing: impact of nutrition and exercise". Acta Physiologica. 216 (1): 15–41. doi:10.1111/apha.12532. PMC 4843955
 . PMID 26010896.
. PMID 26010896. - 1 2 Brioche T, Pagano AF, Py G, Chopard A (April 2016). "Muscle wasting and aging: Experimental models, fatty infiltrations, and prevention". Molecular Aspects of Medicine. doi:10.1016/j.mam.2016.04.006. PMID 27106402.
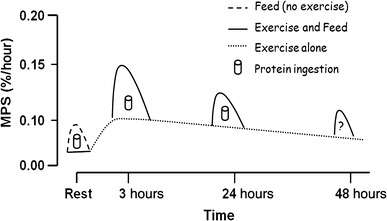
- ↑ Drummond MJ, Dreyer HC, Fry CS, Glynn EL, Rasmussen BB (April 2009). "Nutritional and contractile regulation of human skeletal muscle protein synthesis and mTORC1 signaling". Journal of Applied Physiology. 106 (4): 1374–84. doi:10.1152/japplphysiol.91397.2008. PMC 2698645
 . PMID 19150856.
. PMID 19150856. - ↑ Salto R, Vílchez JD, Girón MD, Cabrera E, Campos N, Manzano M, Rueda R, López-Pedrosa JM (2015). "β-Hydroxy-β-Methylbutyrate (HMB) Promotes Neurite Outgrowth in Neuro2a Cells". PloS One. 10 (8): e0135614. doi:10.1371/journal.pone.0135614. PMC 4534402
 . PMID 26267903.
. PMID 26267903. - ↑ Kougias DG, Nolan SO, Koss WA, Kim T, Hankosky ER, Gulley JM, Juraska JM (April 2016). "Beta-hydroxy-beta-methylbutyrate ameliorates aging effects in the dendritic tree of pyramidal neurons in the medial prefrontal cortex of both male and female rats". Neurobiology of Aging. 40: 78–85. doi:10.1016/j.neurobiolaging.2016.01.004. PMID 26973106.
- 1 2 Phillips SM (May 2014). "A brief review of critical processes in exercise-induced muscular hypertrophy". Sports Med. 44 Suppl 1: S71–S77. doi:10.1007/s40279-014-0152-3. PMC 4008813
 . PMID 24791918.
. PMID 24791918. - ↑ Jimenez SA, Cronin PM, Koenig AS, O'Brien MS, Castro SV (15 February 2012). Varga J, Talavera F, Goldberg E, Mechaber AJ, Diamond HS, eds. "Scleroderma". Medscape Reference. WebMD. Retrieved 5 March 2014.
- ↑ Hajj-ali RA (June 2013). "Systemic Sclerosis". Merck Manual Professional. Merck Sharp & Dohme Corp. Retrieved 5 March 2014.
- ↑ Zhou H, Luo Y, Huang S (September 2010). "Updates of mTOR inhibitors". Anti-Cancer Agents in Medicinal Chemistry. 10 (7): 571–81. doi:10.2174/187152010793498663. PMC 2980558
 . PMID 20812900.
. PMID 20812900. - ↑ Faivre S, Kroemer G, Raymond E (August 2006). "Current development of mTOR inhibitors as anticancer agents". Nature Reviews. Drug Discovery. 5 (8): 671–88. doi:10.1038/nrd2062. PMID 16883305.
- ↑ Hasty P (February 2010). "Rapamycin: the cure for all that ails". Journal of Molecular Cell Biology. 2 (1): 17–9. doi:10.1093/jmcb/mjp033. PMID 19805415.
- ↑ Bové J, Martínez-Vicente M, Vila M (August 2011). "Fighting neurodegeneration with rapamycin: mechanistic insights". Nature Reviews. Neuroscience. 12 (8): 437–52. doi:10.1038/nrn3068. PMID 21772323.
- ↑ "mTOR protein interactors". Human Protein Reference Database. Johns Hopkins University and the Institute of Bioinformatics. Retrieved 2010-12-06.
- ↑ Kumar V, Sabatini D, Pandey P, Gingras AC, Majumder PK, Kumar M, Yuan ZM, Carmichael G, Weichselbaum R, Sonenberg N, Kufe D, Kharbanda S (April 2000). "Regulation of the rapamycin and FKBP-target 1/mammalian target of rapamycin and cap-dependent initiation of translation by the c-Abl protein-tyrosine kinase". The Journal of Biological Chemistry. 275 (15): 10779–87. doi:10.1074/jbc.275.15.10779. PMID 10753870.
- ↑ Sekulić A, Hudson CC, Homme JL, Yin P, Otterness DM, Karnitz LM, Abraham RT (July 2000). "A direct linkage between the phosphoinositide 3-kinase-AKT signaling pathway and the mammalian target of rapamycin in mitogen-stimulated and transformed cells". Cancer Research. 60 (13): 3504–13. PMID 10910062.
- ↑ Cheng SW, Fryer LG, Carling D, Shepherd PR (April 2004). "Thr2446 is a novel mammalian target of rapamycin (mTOR) phosphorylation site regulated by nutrient status". The Journal of Biological Chemistry. 279 (16): 15719–22. doi:10.1074/jbc.C300534200. PMID 14970221.
- ↑ Choi JH, Bertram PG, Drenan R, Carvalho J, Zhou HH, Zheng XF (October 2002). "The FKBP12-rapamycin-associated protein (FRAP) is a CLIP-170 kinase". EMBO Reports. 3 (10): 988–94. doi:10.1093/embo-reports/kvf197. PMC 1307618
 . PMID 12231510.
. PMID 12231510. - ↑ Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC (April 2006). "mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin". The EMBO Journal. 25 (8): 1659–68. doi:10.1038/sj.emboj.7601047. PMC 1440840
 . PMID 16541103.
. PMID 16541103. - 1 2 Schalm SS, Fingar DC, Sabatini DM, Blenis J (May 2003). "TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function". Current Biology. 13 (10): 797–806. doi:10.1016/S0960-9822(03)00329-4. PMID 12747827.
- 1 2 3 Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K (July 2002). "Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action". Cell. 110 (2): 177–89. doi:10.1016/S0092-8674(02)00833-4. PMID 12150926.
- 1 2 Wang L, Rhodes CJ, Lawrence JC (August 2006). "Activation of mammalian target of rapamycin (mTOR) by insulin is associated with stimulation of 4EBP1 binding to dimeric mTOR complex 1". The Journal of Biological Chemistry. 281 (34): 24293–303. doi:10.1074/jbc.M603566200. PMID 16798736.
- 1 2 3 Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J (April 2005). "Rheb binds and regulates the mTOR kinase". Current Biology. 15 (8): 702–13. doi:10.1016/j.cub.2005.02.053. PMID 15854902.
- 1 2 Takahashi T, Hara K, Inoue H, Kawa Y, Tokunaga C, Hidayat S, Yoshino K, Kuroda Y, Yonezawa K (September 2000). "Carboxyl-terminal region conserved among phosphoinositide-kinase-related kinases is indispensable for mTOR function in vivo and in vitro". Genes to Cells. 5 (9): 765–75. doi:10.1046/j.1365-2443.2000.00365.x. PMID 10971657.
- 1 2 Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM (February 1998). "RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1". Proceedings of the National Academy of Sciences of the United States of America. 95 (4): 1432–7. doi:10.1073/pnas.95.4.1432. PMC 19032
 . PMID 9465032.
. PMID 9465032. - ↑ Wang X, Beugnet A, Murakami M, Yamanaka S, Proud CG (April 2005). "Distinct signaling events downstream of mTOR cooperate to mediate the effects of amino acids and insulin on initiation factor 4E-binding proteins". Molecular and Cellular Biology. 25 (7): 2558–72. doi:10.1128/MCB.25.7.2558-2572.2005. PMC 1061630
 . PMID 15767663.
. PMID 15767663. - ↑ Choi J, Chen J, Schreiber SL, Clardy J (July 1996). "Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP". Science. 273 (5272): 239–42. doi:10.1126/science.273.5272.239. PMID 8662507.
- ↑ Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D (August 2004). "Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals". Proceedings of the National Academy of Sciences of the United States of America. 101 (33): 12288–93. doi:10.1073/pnas.0404041101. PMC 514471
 . PMID 15284440.
. PMID 15284440. - ↑ Banaszynski LA, Liu CW, Wandless TJ (April 2005). "Characterization of the FKBP.rapamycin.FRB ternary complex". Journal of the American Chemical Society. 127 (13): 4715–21. doi:10.1021/ja043277y. PMID 15796538.
- ↑ Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT (January 1995). "Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells". The Journal of Biological Chemistry. 270 (2): 815–22. doi:10.1074/jbc.270.2.815. PMID 7822316.
- ↑ Sabatini DM, Barrow RK, Blackshaw S, Burnett PE, Lai MM, Field ME, Bahr BA, Kirsch J, Betz H, Snyder SH (May 1999). "Interaction of RAFT1 with gephyrin required for rapamycin-sensitive signaling". Science. 284 (5417): 1161–4. doi:10.1126/science.284.5417.1161. PMID 10325225.
- ↑ Ha SH, Kim DH, Kim IS, Kim JH, Lee MN, Lee HJ, Kim JH, Jang SK, Suh PG, Ryu SH (December 2006). "PLD2 forms a functional complex with mTOR/raptor to transduce mitogenic signals". Cellular Signalling. 18 (12): 2283–91. doi:10.1016/j.cellsig.2006.05.021. PMID 16837165.
- ↑ Buerger C, DeVries B, Stambolic V (June 2006). "Localization of Rheb to the endomembrane is critical for its signaling function". Biochemical and Biophysical Research Communications. 344 (3): 869–80. doi:10.1016/j.bbrc.2006.03.220. PMID 16631613.
- 1 2 Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B (October 2006). "SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity". Cell. 127 (1): 125–37. doi:10.1016/j.cell.2006.08.033. PMID 16962653.
- ↑ McMahon LP, Yue W, Santen RJ, Lawrence JC (January 2005). "Farnesylthiosalicylic acid inhibits mammalian target of rapamycin (mTOR) activity both in cells and in vitro by promoting dissociation of the mTOR-raptor complex". Molecular Endocrinology. 19 (1): 175–83. doi:10.1210/me.2004-0305. PMID 15459249.
- ↑ Oshiro N, Yoshino K, Hidayat S, Tokunaga C, Hara K, Eguchi S, Avruch J, Yonezawa K (April 2004). "Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function". Genes to Cells. 9 (4): 359–66. doi:10.1111/j.1356-9597.2004.00727.x. PMID 15066126.
- ↑ Kawai S, Enzan H, Hayashi Y, Jin YL, Guo LM, Miyazaki E, Toi M, Kuroda N, Hiroi M, Saibara T, Nakayama H (July 2003). "Vinculin: a novel marker for quiescent and activated hepatic stellate cells in human and rat livers". Virchows Archiv. 443 (1): 78–86. doi:10.1007/s00428-003-0804-4. PMID 12719976.
- ↑ Choi KM, McMahon LP, Lawrence JC (May 2003). "Two motifs in the translational repressor PHAS-I required for efficient phosphorylation by mammalian target of rapamycin and for recognition by raptor". The Journal of Biological Chemistry. 278 (22): 19667–73. doi:10.1074/jbc.M301142200. PMID 12665511.
- 1 2 Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K (May 2003). "The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif". The Journal of Biological Chemistry. 278 (18): 15461–4. doi:10.1074/jbc.C200665200. PMID 12604610.
- 1 2 Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM (April 2006). "Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB". Molecular Cell. 22 (2): 159–68. doi:10.1016/j.molcel.2006.03.029. PMID 16603397.
- ↑ Tzatsos A, Kandror KV (January 2006). "Nutrients suppress phosphatidylinositol 3-kinase/Akt signaling via raptor-dependent mTOR-mediated insulin receptor substrate 1 phosphorylation". Molecular and Cellular Biology. 26 (1): 63–76. doi:10.1128/MCB.26.1.63-76.2006. PMC 1317643
 . PMID 16354680.
. PMID 16354680. - 1 2 3 Sarbassov DD, Sabatini DM (November 2005). "Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex". The Journal of Biological Chemistry. 280 (47): 39505–9. doi:10.1074/jbc.M506096200. PMID 16183647.
- 1 2 Yang Q, Inoki K, Ikenoue T, Guan KL (October 2006). "Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity". Genes & Development. 20 (20): 2820–32. doi:10.1101/gad.1461206. PMC 1619946
 . PMID 17043309.
. PMID 17043309. - ↑ Kumar V, Pandey P, Sabatini D, Kumar M, Majumder PK, Bharti A, Carmichael G, Kufe D, Kharbanda S (March 2000). "Functional interaction between RAFT1/FRAP/mTOR and protein kinase cdelta in the regulation of cap-dependent initiation of translation". The EMBO Journal. 19 (5): 1087–97. doi:10.1093/emboj/19.5.1087. PMC 305647
 . PMID 10698949.
. PMID 10698949. - ↑ Long X, Ortiz-Vega S, Lin Y, Avruch J (June 2005). "Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency". The Journal of Biological Chemistry. 280 (25): 23433–6. doi:10.1074/jbc.C500169200. PMID 15878852.
- ↑ Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG (May 2005). "The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses". The Journal of Biological Chemistry. 280 (19): 18717–27. doi:10.1074/jbc.M414499200. PMID 15772076.
- ↑ Bernardi R, Guernah I, Jin D, Grisendi S, Alimonti A, Teruya-Feldstein J, Cordon-Cardo C, Simon MC, Rafii S, Pandolfi PP (August 2006). "PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR". Nature. 442 (7104): 779–85. doi:10.1038/nature05029. PMID 16915281.
- ↑ Saitoh M, Pullen N, Brennan P, Cantrell D, Dennis PB, Thomas G (May 2002). "Regulation of an activated S6 kinase 1 variant reveals a novel mammalian target of rapamycin phosphorylation site". The Journal of Biological Chemistry. 277 (22): 20104–12. doi:10.1074/jbc.M201745200. PMID 11914378.
- ↑ Chiang GG, Abraham RT (July 2005). "Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase". The Journal of Biological Chemistry. 280 (27): 25485–90. doi:10.1074/jbc.M501707200. PMID 15899889.
- ↑ Holz MK, Blenis J (July 2005). "Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase". The Journal of Biological Chemistry. 280 (28): 26089–93. doi:10.1074/jbc.M504045200. PMID 15905173.
- ↑ Isotani S, Hara K, Tokunaga C, Inoue H, Avruch J, Yonezawa K (November 1999). "Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro". The Journal of Biological Chemistry. 274 (48): 34493–8. doi:10.1074/jbc.274.48.34493. PMID 10567431.
- ↑ Toral-Barza L, Zhang WG, Lamison C, Larocque J, Gibbons J, Yu K (June 2005). "Characterization of the cloned full-length and a truncated human target of rapamycin: activity, specificity, and enzyme inhibition as studied by a high capacity assay". Biochemical and Biophysical Research Communications. 332 (1): 304–10. doi:10.1016/j.bbrc.2005.04.117. PMID 15896331.
- 1 2 Ali SM, Sabatini DM (May 2005). "Structure of S6 kinase 1 determines whether raptor-mTOR or rictor-mTOR phosphorylates its hydrophobic motif site". The Journal of Biological Chemistry. 280 (20): 19445–8. doi:10.1074/jbc.C500125200. PMID 15809305.
- ↑ Edinger AL, Linardic CM, Chiang GG, Thompson CB, Abraham RT (December 2003). "Differential effects of rapamycin on mammalian target of rapamycin signaling functions in mammalian cells". Cancer Research. 63 (23): 8451–60. PMID 14679009.
- ↑ Leone M, Crowell KJ, Chen J, Jung D, Chiang GG, Sareth S, Abraham RT, Pellecchia M (August 2006). "The FRB domain of mTOR: NMR solution structure and inhibitor design". Biochemistry. 45 (34): 10294–302. doi:10.1021/bi060976+. PMID 16922504.
- ↑ Kristof AS, Marks-Konczalik J, Billings E, Moss J (September 2003). "Stimulation of signal transducer and activator of transcription-1 (STAT1)-dependent gene transcription by lipopolysaccharide and interferon-gamma is regulated by mammalian target of rapamycin". The Journal of Biological Chemistry. 278 (36): 33637–44. doi:10.1074/jbc.M301053200. PMID 12807916.
- ↑ Yokogami K, Wakisaka S, Avruch J, Reeves SA (January 2000). "Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR". Current Biology. 10 (1): 47–50. doi:10.1016/S0960-9822(99)00268-7. PMID 10660304.
- ↑ Kusaba H, Ghosh P, Derin R, Buchholz M, Sasaki C, Madara K, Longo DL (January 2005). "Interleukin-12-induced interferon-gamma production by human peripheral blood T cells is regulated by mammalian target of rapamycin (mTOR)". The Journal of Biological Chemistry. 280 (2): 1037–43. doi:10.1074/jbc.M405204200. PMID 15522880.
- ↑ Cang C, Zhou Y, Navarro B, Seo YJ, Aranda K, Shi L, Battaglia-Hsu S, Nissim I, Clapham DE, Ren D (February 2013). "mTOR regulates lysosomal ATP-sensitive two-pore Na(+) channels to adapt to metabolic state". Cell. 152 (4): 778–90. doi:10.1016/j.cell.2013.01.023. PMC 3908667
 . PMID 23394946.
. PMID 23394946. - ↑ Wu S, Mikhailov A, Kallo-Hosein H, Hara K, Yonezawa K, Avruch J (January 2002). "Characterization of ubiquilin 1, an mTOR-interacting protein". Biochimica et Biophysica Acta. 1542 (1-3): 41–56. doi:10.1016/S0167-4889(01)00164-1. PMID 11853878.
External links
- mTOR protein at the US National Library of Medicine Medical Subject Headings (MeSH)
- "mTOR Signaling Pathway in Pathway Interaction Database". National Cancer Institute.