K562 cells
K562 cells were the first human immortalised myelogenous leukemia line to be established. K562 cells are of the erythroleukemia type, and the line is derived from a 53-year-old female chronic myelogenous leukemia patient in blast crisis.[1][2] The cells are non-adherent and rounded, are positive for the bcr:abl fusion gene, and bear some proteomic resemblance to both undifferentiated granulocytes[3] and erythrocytes.[4]
In culture they exhibit much less clumping than many other suspension lines, presumably due to the downregulation of surface adhesion molecules by bcr:abl.[5] However, another study suggests that bcr:abl over-expression may actually increase cell adherence to cell culture plastic.[6] K562 cells can spontaneously develop characteristics similar to early-stage erythrocytes, granulocytes and monocytes[7] and are easily killed by natural killer cells[8] as they lack the MHC complex required to inhibit NK activity.[2] They also lack any trace of Epstein-Barr virus and other herpesviruses. In addition to the Philadelphia chromosome they also exhibit a second reciprocal translocation between the long arm of chromosome 15 with chromosome 17.[1]
Two sub-lines are available which express MHC class-I A2[9] and A3.[10]
K562 Cell Cycle and Regulation
Many factors and components play a role in the cell cycle of K562 cells in terms of growth, cell differentiation, and apoptosis.[11] The growth of these leukemic cells are controlled by either initiating cell differentiation or apoptosis to occur.[12]
Cell differentiation is induced by the deacytylase activity in these “undifferentiated progenitor cells”, which alters the phenotype and morphology of the K562 cells.[11] The change in phenotype induces a decrease in the growth rate and leads the K562 cells to the terminal path of becoming mature erythroids, monocytes, and mature macrophages.[11] These changes can also drive the leukemic cells to a state of stress, which allows for increased sensitivity of the cells to drugs that initiate apoptosis.[11]
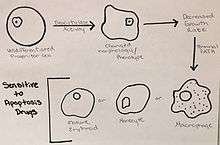
The problem with K562 cells, and many other cancer cell types, is that there is an overabundance of Aurora kinases.[13] These kinases play a role in the formation of spindles, separation of chromosomes, as well as cytokinesis.[13] These functions are necessary in cells in order to divide and regenerate tissues, and play a maintenance role in homeostatic functions, but the overabundance of Aurora kinases allows for uncontrolled cellular division, resulting in cancer.[13] Inhibiting these is an important regulation mechanism of cancer, because it prevents cells from moving into mitosis.[13]

Apoptosis is an important mechanism in regulating K562 cells and can be induced by the changes in the metabolic state of the cells.[11] There are many different cellular components involved in the cycle of apoptosis such as, BCR/ABL, Bcl-2, Bax protein, and Cytochrome C.[12] The tumor suppressor gene, p53, is also important in the cell cycle regulation of K562 cells.[14] This gene targets the cyclin-dependent kinase inhibitor, p21, and causes cell differentiation, cell cycle arrest in G1, and ultimately apoptosis.[15] When the levels of these components are thrown off, they can either no longer inhibit apoptosis of the cancer cells, like BCR/ABL, or they cause apoptosis to be induced, such as Bax and cytochrome C.[12] These components are key in the mitochondria, and due to this, it has been supported that apoptosis uses the mitochondrial apoptosis pathway.[12] The offset of these cellular components from their balance point causes morphological changes, which result in the K562 cells being arrested in the G2/M phase of the cell cycle.[12] This arrest leads to “shrinkage, blebbing, nuclear fragmentation, chromatin condensing” and other morphological changes that cause the cell to program death at this point.[12]
The ability to induce these changes in K562 cell cycle and cell cycle regulation provides targets for cancer drugs.[16] One of these drugs is Imatinib, which inhibits BCR/ABL causing growth to seize and apoptosis to begin.[17] Another important group of regulators of the K562 line are Sirtuins, referred to as SIRTS.[11] These play a role in cellular stress, metabolism, and autophagy, by interacting with deacytylases activity in the cell.[11] Other methods being focused on in the regulation of K562 cells include therapeutic methods like Polyphylin D, which caused differentiation from the progenitor state to occur, and for apoptosis to begin.[12] In addition to this method, Chinese medicine using the Eriocaulon seiboldianum plant is being researched and displaying a role in the inhibition of Aurora kinases.[13]
The research with the E. seiboldianum edible Chinese medicine plant is quite interesting. It has been supported that this medical plant helps regulate K562 cell growth by inhibiting Aurora kinase activity, up regulating apoptosis proteins such as p53 and Bax, as well as reducing the levels of the Bcl-2 anti-apoptosis protein.[18] These changes and the imbalance in the protein levels within the K562 cells cause spindle defects and arrests the cell in the G2-M phase of the cell cycle, ultimately causing the cell to undergo apoptosis.[18]
External links
References
- 1 2 Lozzio, C.B.; Lozzio, B.B. (1975), "Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome", Blood, 45 (3): 321–34, PMID 163658
- 1 2 Drexler, H.G. (2000), The Leukemia-Lymphoma Cell Line Factsbook, San Diego: Academic Press
- ↑ Klein, E.; Ben-Bassat, H.; Neumann, H.; Ralph, P.; Zeuthen, J.; Polliack, A.; Vánky, F. (1976), "Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia", International Journal of Cancer, 18 (4): 421–31, doi:10.1002/ijc.2910180405, PMID 789258
- ↑ Andersson, L.C.; Nilsson, K.; Gahmberg, C.G. (1979), "K562 - A human erythroleukemic cell line", International Journal of Cancer, 23 (2): 143–7, doi:10.1002/ijc.2910230202, PMID 367973
- ↑ Jongen-Lavrencic, M (2005). "BCR/ABL-mediated downregulation of genes implicated in cell adhesion and motility leads to impaired migration toward CCR7 ligands CCL19 and CCL21 in primary BCR/ABL-positive cells". Leukemia. 19 (3): 373–380. doi:10.1038/sj.leu.2403626. PMID 15674360.
- ↑ Karimiani, EG; Marriage, F; Merritt, AJ; Burthem, J; Byers, RJ; Day, PJ (Mar 2014). "Single-cell analysis of K562 cells: an imatinib-resistant subpopulation is adherent and has upregulated expression of BCR-ABL mRNA and protein.". Experimental hematology. 42 (3): 183–191.e5. doi:10.1016/j.exphem.2013.11.006. PMID 24269846.
- ↑ Lozzio, B.B.; Lozzio, C.B.; Bamberger, E.G.; Feliu, A.S. (1981), "A multipotential leukemia cell line (K-562) of human origin", Proceedings of the Society for Experimental Biology and Medicine, 166 (4): 546–50, doi:10.3181/00379727-166-41106, PMID 7194480
- ↑ Lozzio, B.B.; Lozzio, C.B. (1979), "Properties and usefulness of the original K-562 human myelogenous leukemia cell line", Leukemia Research, 3 (6): 363–70, doi:10.1016/0145-2126(79)90033-X, PMID 95026
- ↑ Britten, C.M.; Meyer, R.G.; Kreer, T.; Drexler, I.; Wölfel, T.; Herr, W. (2002), "The use of HLA-A*0201-transfected K562 as standard antigen-presenting cells for CD8(+) T lymphocytes in IFN-gamma ELISPOT assays", Journal of Immunological Methods, 259 (1–2): 95–110, doi:10.1016/S0022-1759(01)00499-9, PMID 11730845
- ↑ Clark, R.E.; Dodi, I.A.; Hill, S.C.; Lill, J.R.; Aubert, G.; Macintyre, A.R.; Rojas, J.; Bourdon, A.; et al. (2001), "Direct evidence that leukemic cells present HLA-associated immunogenic peptides derived from the BCR-ABL b3a2 fusion protein", Blood, 98 (10): 2887–93, doi:10.1182/blood.V98.10.2887, PMID 11698267
- 1 2 3 4 5 6 7 Duncan, Mark; DeLuca, Teresa; Kuo, Hsin-Yu; Yi, Minchang; Mrksich, Milan; Miller, William (2016). "SIRT1 is a critical regulator of K562 cell growth, survival, and differentiation". Experimental Cell Research. 344: 40–52. doi:10.1016/j.yexcr.2016.04.010.
- 1 2 3 4 5 6 7 Yang, Chunhui; Cai, Hong; Meng, Xiuxiang (2016). "Polyphyllin D induces apoptosis and differentiation in K562 human leukemia cells". Internal Immunopharmacology. 36: 17–22. doi:10.1016/j.intimp.2016.04.011.
- 1 2 3 4 5 Fan, Yanhua; Lu, Hongyuan; An, Li; Wang, Changli; Zhou, Zhipeng; Feng, Fan; Ma, Hongda; Xu, Yongnan; Zhao, Qingchun (2016). "Effect of active faction of Eriocaulon sieboldianum on human leukemia K563 cells via proliferation inhibition, cell cycle arrest and apoptosis induction". Environmental Toxicology and Pharmacology. 43: 13–20. doi:10.1016/j.etap.2015.11.001.
- ↑ Chylicki, K; Ehinger, M; Svedberg, H; Bergh, G; Olsson, I; Gullberg, U (2000). "p53 mediated differentiation of the erythroleukemia cell line K562". Cell Growth & Differentiation: the Molecular Biology Journal of the American Association for Cancer Research. 11 (6): 315–324. PMID 10910098.
- ↑ Chylicki, K; Ehinger, M; Svedberg, H; Bergh, G; Olsson, I; Gullberg, U (2000). "p53 mediated differentiation of the erythroleukemia cell line K562". Cell Growth & Differentiation: the Molecular Biology Journal of the American Association for Cancer Research. 11 (6): 315–324. PMID 10910098.
- ↑ Wang, Jian; Li, Qinghua; Wang, Chijuan; Xiong, Q; Lin, Y; Sun, Q; Jin, H; Yang, F; Ren, X; Pang, T (2016). "Knock-down of CIAPIN1 sensitizes K562 chronic myeloid leukemia cells to Imatinib by regulation of cell cycle and apoptosis-associated members via NF-KB and ERK5 signaling pathway". Biochemical Pharmacology. 99: 132–145. doi:10.1016/j.bcp.2015.12.002.
- ↑ Wang, Jian; Li, Qinghua; Wang, Chijuan; Xiong, Q; Lin, Y; Sun, Q; Jin, H; Yang, F; Ren, X; Pang, T (2016). "Knock-down of CIAPIN1 sensitizes K562 chronic myeloid leukemia cells to Imatinib by regulation of cell cycle and apoptosis-associated members via NF-KB and ERK5 signaling pathway". Biochemical Pharmacology. 99: 132–145. doi:10.1016/j.bcp.2015.12.002.
- 1 2 Fan, Yanhua (2016). "Effect of activation fraction of Eriocaulon sieboldianum on human leukemia K562 cells via proliferation inhibition, cell cycle arrest and apoptosis induction". Environmental Toxicology and Pharmacology. 43: 13–20.