Heparan sulfate
Heparan sulfate (HS) is a linear polysaccharide found in all animal tissues. It occurs as a proteoglycan (HSPG) in which two or three HS chains are attached in close proximity to cell surface or extracellular matrix proteins.[1][2] It is in this form that HS binds to a variety of protein ligands and regulates a wide variety of biological activities, including developmental processes, angiogenesis, blood coagulation, abolishing detachment activity by GrB (Granzyme B),[3] and tumour metastasis. HS has been shown to serve as cellular receptor for a number of viruses including the respiratory syncytial virus (Hallak et al. 2000)
Proteoglycans
The major cell membrane HSPGs are the transmembrane syndecans and the glycosylphosphatidylinositol (GPI) anchored glypicans. Other minor forms of membrane HSPG include betaglycan[4] and the V-3 isoform of CD44 present on keratinocytes and activated monocytes.[5]
In the extracellular matrix, especially basement membranes, the multi-domain perlecan, agrin and collagen XVIII core proteins are the main HS-bearing species.
Possible role in Alzheimer's disease and Parkinson's disease
HSPGs have been implicated in the progression of neurodegenerative diseases. Tau fibrils bind to cell surface HSPGs in a similar manner to prions, triggering intracellular fibril aggregation, suggesting a unifying mechanism for tauopathies and synucleopathies, and potential drug targets.[6]
Structure and differences from heparin
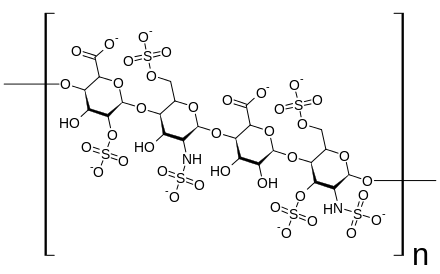
Heparan sulfate is a member of the glycosaminoglycan family of carbohydrates and is very closely related in structure to heparin. Both consist of a variably sulfated repeating disaccharide unit. The main disaccharide units that occur in heparan sulfate and heparin are shown below.
The most common disaccharide unit within heparan sulfate is composed of a glucuronic acid (GlcA) linked to N-acetylglucosamine (GlcNAc) typically making up around 50% of the total disaccharide units. Compare this to heparin where IdoA(2S)-GlcNS(6S) makes up 85% of heparins from beef lung and about 75% of those from porcine intestinal mucosa. Problems arise when defining hybrid GAGs that contain both 'heparin-like' and 'HS-like' structures. It has been suggested that a GAG should qualify as heparin only if its content of N-sulfate groups largely exceeds that of N-acetyl groups and the concentration of O-sulfate groups exceeds those of N-sulfate.[7]
Not shown below are the rare disaccharides containing a 3-O-sulfated glucosamine (GlcNS(3S,6S) or a free amine group (GlcNH3+). Under physiological conditions the ester and amide sulfate groups are deprotonated and attract positively charged counterions to form a salt. It is in this form that HS is thought to exist at the cell surface.
-
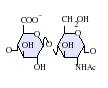
GlcA-GlcNAc -
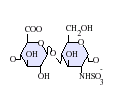
GlcA-GlcNS -
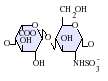
IdoA-GlcNS -
-GlcNS.png)
IdoA(2S)-GlcNS -
.png)
IdoA-GlcNS(6S) -
-GlcNS(6S).png)
IdoA(2S)-GlcNS(6S)
Abbreviations
- GlcA = β-D-glucuronic acid
- IdoA = α-L-iduronic acid
- IdoA(2S) = 2-O-sulfo-α-L-iduronic acid
- GlcNAc = 2-deoxy-2-acetamido-α-D-glucopyranosyl
- GlcNS = 2-deoxy-2-sulfamido-α-D-glucopyranosyl
- GlcNS(6S) = 2-deoxy-2-sulfamido-α-D-glucopyranosyl-6-O-sulfate
Biosynthesis
Many different cell types produce HS chains with many different primary structures. Therefore, there is a great deal of variability in the way HS chains are synthesised. However, essential to the formation of HS regardless of primary sequence is a range of biosynthetic enzymes. These enzymes consist of multiple glycosyltransferases, sulfotransferases and an epimerase. These same enzymes also synthesize heparin.
In the 1980s, Jeffrey Esko was the first to isolate and characterize animal cell mutants altered in the assembly of heparan sulfate.[8] Many of these enzymes have now been purified, molecularly cloned and their expression patterns studied. From this and early work on the fundamental stages of HS/heparin biosynthesis using a mouse mastocytoma cell free system a lot is known about the order of enzyme reactions and specificity.[9]
Chain initiation
HS synthesis initiates with the transfer of xylose from UDP-xylose by xylosyltransferase (XT) to specific serine residues within the protein core. Attachment of two galactose (Gal) residues by galactosyltransferases I and II (GalTI and GalTII) and glucuronic acid (GlcA) by glucuronosyltransferase I (GlcATI) completes the formation of a core protein linkage tetrasaccharide
βGlcA-1,3-βGal-1,3-βGal-1,4-βXyl.
Xylose attachment to the core protein is thought to occur in the endoplasmic reticulum (ER) with further assembly of the linkage region and the remainder of the chain occurring in the golgi apparatus.
The pathways for HS/heparin or chondroitin sulfate (CS) and dermatan sulfate (DS) biosynthesis diverge after the formation of this common linkage structure. The next enzyme to act, GlcNAcT-I or GalNAcT-I, directs synthesis, either to HS/heparin or CS/DS, respectively.
Chain elongation
After attachment of the first N-acetylglucosamine (GlcNAc) residue elongation of the tetrasacchride linker is continued by the stepwise addition of GlcA and GlcNAc residues. These are transferred from their respective UDP-sugar nucleotides. This is carried out by one or more related enzymes whose genes are members of the exostoses (EXT) gene family of tumour suppressors.
Mutations at the EXT1-3 gene loci in humans leads to an inability of cells to produce HS and to the development of the disease Multiple Hereditary Exostoses (MHE). MHE is characterized by cartilage-capped tumours, known as osteochondromas or exostoses, which develop primarily on the long bones of affected individuals from early childhood until puberty.
Chain modification
As the chain polymerises, it undergoes a series of modification reactions carried out by four classes of sulfotransferases and an epimerase. The availability of the sulfate donor PAPS is crucial to the activity of the sulfotransferases.[10][11]
N-deacetylation/N-sulfation
The first polymer modification is the N-deacetylation/N-sulfation of GlcNAc residues into GlcNS. This is a prerequisite for all subsequent modification reactions, and is carried out by one or more members of a family of four GlcNAc N-deacetylase/N-sulfotransferase enzymes (NDSTs). In early studies, it was shown that modifying enzymes could recognize and act on any N-acetylated residue in the forming polymer.[12] Therefore, the modification of GlcNAc residues should occur randomly throughout the chain. However, in HS, N-sulfated residues are mainly grouped together and separated by regions of N-acetylation where GlcNAc remains unmodified.
Generation of GlcNH2
Due to the N-deacetylase and N-sulfotransferase being carried out by the same enzyme N-sulfation is normally tightly coupled to N-acetylation. GlcNH2 residues resulting from apparent uncoupling of the two activities have been found in heparin and some species of HS.[13]
Epimerisation and 2-O-sulfation
Epimerisation is catalysed by one enzyme, the GlcA C5 epimerase or heparosan-N-sulfate-glucuronate 5-epimerase (EC 5.1.3.17). This enzyme epimerises GlcA to iduronic acid (IdoA). Substrate recognition requires that the GlcN residue linked to the non-reducing side of a potential GlcA target be N-sulfated. Uronosyl-2-O-sulfotransferase (2OST) sulfates the resulting IdoA residues.
6-O-sulfation
Three glucosaminyl 6-O-transferases (6OSTs) have been identified that result in the formation of GlcNS(6S) adjacent to sulfated or non-sulfated IdoA. GlcNAc(6S) is also found in mature HS chains.
3-O-sulfation
Currently seven glucosaminyl 3-O-sulfotransferases (3OSTs) are known to exist in mammals (eight in zebrafish).[14][15] The 3OST enzymes create a number of possible 3-O-sulfated disaccharides, including GlcA-GlcNS(3S±6S) (modified by 3OST1 and 3OST5), IdoA(2S)-GlcNH2(3S±6S)(modified by 3OST3a, 3OST3b, 3OST5 and 3OST6) and GlcA/IdoA(2S)-GlcNS(3S) (modified by 3OST2 and 3OST4).[16][17][18][19] As with all other HS sulfotransferases, the 3OSTs use 3'-phosphoadenosine-5'-phosphosulfate (PAPS) as a sulfate donor. Despite being the largest family of HS modification enzymes, the 3OSTs produce the rarest HS modification, the 3-O-sulfation of specific glucosamine residues at the C3-OH moiety.[20]
The 3OSTs are divided into two functional subcategories, those that generate an antithrombin III binding site (3OST1 and 3OST5) and those that generate a herpes simplex virus 1 glycoprotein D (HSV-1 gD) binding site (3OST2, 3OST3a, 3OST3b, 3OST4, 3OST5 and 3OST6).[16][17][18][19][21][22][23][24][25][26][27] As the 3OSTs are the largest family of HS modification enzymes and their actions are rate-limiting, substrate specific and produce rare modifications, it has been hypothesized that 3OST modified HS plays an important regulatory role in biological processes.[19][22][22]
Ligand binding
Heparan sulfate binds with a large number of extracellular proteins. These are often collectively called the “heparin interactome” or "heparin-binding proteins", because they are isolated by affinity chromatography on the related polysaccharide heparin, though the terms “heparan sulfate interactome” or "heparanome" are more correct. The functions of heparan sulfate binding proteins ranges from extracellular matrix components, to enzymes and coagulation factors, and most growth factors, cytokines, chemokines and morphogens [28]
Interferon-γ
The cell surface receptor binding region of Interferon-γ overlaps with the HS binding region, near the protein's C-terminal. Binding of HS blocks the receptor binding site and as a result, protein-HS complexes are inactive.[29]
The HS-binding properties of a number of other proteins are also being studied:
- Antithrombin III
- Fibroblast Growth Factors
- Hepatocyte Growth Factor
- Interleukin-8
- Vascular Endothelial Growth Factor
- Wnt/Wingless
- Endostatin
Heparan sulfate analogue
Heparan sulfate analogues are thought to display identical properties as heparan sulfate with exception of being stable in a proteolytic environment like a wound.[30][31] Because heparan sulfate is broken down in chronic wounds by heparanase, the analogues only bind sites where natural heparan sulfate is absent and cannot be broken down by any known heparanases and glycanases.[32] Also the function of the heparan sulfate analogues is the same as heparan sulfate, protecting a variety of protein ligands such as growth factors and cytokines. By holding them in place, the tissue can then use the different protein ligands for proliferation.
References
- ↑ Gallagher, J.T., Lyon, M. (2000). "Molecular structure of Heparan Sulfate and interactions with growth factors and morphogens". In Iozzo, M, V. Proteoglycans: structure, biology and molecular interactions. Marcel Dekker Inc. New York, New York. pp. 27–59.
- ↑ Iozzo, R. V. (1998). "Matrix proteoglycans: from molecular design to cellular function". Annu. Rev. Biochem. 67: 609–652. doi:10.1146/annurev.biochem.67.1.609. PMID 9759499.
- ↑ Buzza, Marguerite; Zamurs, Laura; Sun, Jiuru; Bird, Catherina (June 2005). "Extracellular Matrix Remodeling by Human Granzyme B via Cleavage of Vitronectin, Fibronectin, and Laminin". Journal of Biological Chemistry. 280 (25): 23549–23558. doi:10.1074/jbc.M412001200.
- ↑ Andres, J. L.; Noda, M; et al. (1992). "Binding of two growth factor families to separate domains of the proteoglycan betaglycan". J. Biol. Chem. 267 (9): 5927–30. PMID 1556106.
- ↑ Jackson, D. G.; Dickinson, R; et al. (1995). "Proteoglycan forms of lymphocyte homing receptor CD44 are alternatively spliced variants containing the V-3 exon". J. Cell. Biol. 128 (4): 673–685. doi:10.1083/jcb.128.4.673. PMC 2199896
 . PMID 7532175.
. PMID 7532175. - ↑ Holmes, BB.; Devos, SL.; Kfoury, N.; Li, M.; Jacks, R.; Yanamandra, K.; Ouidja, MO.; Brodsky, FM.; Marasa, J.; Bagchi, D. P.; Kotzbauer, P. T.; Miller, T. M.; Papy-Garcia, D.; Diamond, M. I. (Aug 2013). "Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds". Proc Natl Acad Sci U S A. 110 (33): E3138–47. doi:10.1073/pnas.1301440110. PMC 3746848
 . PMID 23898162.
. PMID 23898162. - ↑ Gallagher, J. T. Walker, A. (1985). "Molecular distinctions between Heparan Sulphate and Heparin: Analysis of sulphation patterns indicates Heparan Sulphate and Heparin are separate families of N-sulphated polysaccharides". Biochem. J. 230 (3): 665–74. PMC 1152670
 . PMID 2933029.
. PMID 2933029. - ↑ Esko, J.D.; Stewart, T.E.; Taylor, W.H. (1985). "Animal cell mutants defective inglycosaminoglycan biosynthesis". Proc. Natl. Acad. Sci. USA. 85: 3197–3201. PMID 3858816.
- ↑ Lindahl, U.; et al. (1998). "Regulated diversity of Heparan Sulfate". J. Biol. Chem. 273 (39): 24979–24982. doi:10.1074/jbc.273.39.24979. PMID 9737951.
- ↑ Silbert, J. E. (10 November 1967). "Formation of a sulfate glycosaminoglycan with a microsomal preparation from mast cells". J. Biol. Chem. 242 (21): 5146–52. PMID 4228675.
- ↑ Carlsson P; Presto J.; et al. (2008). "Heparin/Heparan Sulfate Biosynthesis: Processive formation of N-sulfated domains". J. Biol. Chem. 283 (29): 20008–20014. doi:10.1074/jbc.M801652200. PMID 18487608.
- ↑ Höök, M.; et al. (1975). "Biosynthesis of heparin. Studies on the microsomal sulfation process". J. Biol. Chem. 250 (15): 6065–71. PMID 807579.
- ↑ Toida, T.; et al. (1 January 1997). "Structural differences and the presence of unsubstituted amino groups in heparan sulphates from different tissues and species". Biochem. J. 322(Pt2) (Pt 2): 499–506. PMC 1218218
 . PMID 9065769.
. PMID 9065769. - ↑ Cadwallader, AB; Yost, HJ (February 2007). "Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: III. 2-O-sulfotransferase and C5-epimerases". Developmental Dynamics. 236 (2): 581–6. doi:10.1002/dvdy.21051. PMID 17195182.
- ↑ Xu, D; Tiwari, V; Xia, G; Clement, C; Shukla, D; Liu, J (Jan 15, 2005). "Characterization of heparan sulphate 3-O-sulphotransferase isoform 6 and its role in assisting the entry of herpes simplex virus type 1". The Biochemical Journal. 385 (Pt 2): 451–9. doi:10.1042/BJ20040908. PMC 1134716
 . PMID 15303968.
. PMID 15303968. - 1 2 Shukla, D.; Liu, J.; Blaiklock, P.; Shworak, N.W.; Bai, X.; Esko, J.D.; Cohen, G.H.; Eisenberg, R.J.; Rosenberg, R.D.; Spear, P.G. (1999). "A Novel Role for 3-O-Sulfated Heparan Sulfate in Herpes Simplex Virus 1 Entry". Cell. 99: 13–22. doi:10.1016/s0092-8674(00)80058-6.
- 1 2 Xia, G.; Chen, J.; Tiwari, V.; Ju, W.; Li, J.P.; Malmstrom, A.; Shukla, D.; Liu, J. (2002). "Heparan Sulfate 3-O-Sulfotransferase Isoform 5 Generates Both an Antithrombin-binding Site and an Entry Receptor for Herpes Simplex Virus, Type 1". The Journal of Biological Chemistry. 277 (40): 37912–37919. doi:10.1074/jbc.m204209200.
- 1 2 Xu, D.; Tiwari, V.; Xia, G.; Clement, C.; Shukla, D.; Liu, J. (2005). "Characterization of heparan sulphate 3-O-sulphotransferase isoform 6 and its role in assisting the entry of herpes simplex virus type 1". Biochemistry Journal. 385: 451–459. doi:10.1042/bj20040908.
- 1 2 3 Lawrence, R.; Yabe, T.; HajMohammadi, S.; Rhodes, J.; McNeely, M.; Liu, J.; Lamperti, E.D.; Toselli, P.A.; Lech, M.; Spear, P.G.; Rosenberg, R.D.; Shworak, N.W. (2007). "The principal neuronal gD-type 3-O-sulfotransferases and their products in the central and peripheral nervous system tissues". Matrix Biology. 26: 442–455. doi:10.1016/j.matbio.2007.03.002. PMC 1993827
 . PMID 17482450.
. PMID 17482450. - ↑ Shworak, N.W.; HajMohammadi, S.; de Agostini, A.I.; Rosenberg, R.D. (2003). "Mice deficient in heparan sulfate 3-O-silfotransferase-1: Normal hemostasis with unexpected perinatal phenotypes". Glycoconjugate Journal. 19 (4-5): 355–361. doi:10.1023/a:1025377206600.
- ↑ Liu, J.; Shworak, N.W.; Fritze, L.M.S.; Edelberg, J.M.; Rosenberg, R.D. (1996). "Purification of Heparan Sulfate D-Glucosaminyl 3-O-Sulfotransferase". The Journal of Biological Chemistry. 271 (43): 27072–27082.
- 1 2 3 Shworak, N.W.; Liu, J.; Fritze, L.M.S.; Schwartz, J.J.; Zhang, L.; Logeart, D.; Rosenberg, R.D. (1997). "Molecular Cloning and Expression of Mouse and Human cDNAs Encoding Heparan Sulfate D-Glucosaminyl 3-O-Sulfotransferase". The Journal of Biological Chemistry. 272 (44): 28008–28019. doi:10.1074/jbc.272.44.28008. PMID 9346953.
- ↑ Shworak, N.W.; Liu, J.; Petros, L.M.; Zhang, L.; Kobayashi, M.; Copeland, N.G.; Jenkins, N.A.; Rosenberg, R.D. (1999). "Multiple Isoforms of Heparan Sulfate D-Glucosaminyl 3-O-Sulfotransferase". The Journal of Biological Chemistry. 274 (8): 5170–5184. doi:10.1074/jbc.274.8.5170. PMID 9988767.
- ↑ Chen, J.; Duncan, M.B.; Carrick, K.; Pope, R.M.; Liu, J. (2003). "Biosynthesis of 3-O-sulfated heparan sulfate: unique substrate specificity of heparan sulfate 3-O-sulfotransferase isoform 5". Glycobiology. 13 (11): 785–794. doi:10.1093/glycob/cwg101.
- ↑ Duncan, M.B.; Chen, J.; Krise, J.P.; Liu, J. (2004). "The biosynthesis of anticoagulant heparan sulfate by the heparan sulfate 3-O-sulfotransferase isoform 5". Biochimica et Biophysica Acta. 1671: 34–43. doi:10.1016/j.bbagen.2003.12.010.
- ↑ Chen, J.; Liu, J. (2005). "Characterization of the structure of antithrombin-binding heparan sulfate generated by heparan sulfate 3-O-sulfotransferase 5". Biochimica et Biophysica Acta. 1725: 190–200. doi:10.1016/j.bbagen.2005.06.012.
- ↑ Girardin, E.P.; HajMohammadi, S.; Birmele, B.; Helisch, A.; Shworak, N.W.; de Agostini, A.I. (2005). "Synthesis of Anticoagulantly Active Heparan Sulfate Proteoglycans by Glomerular Epithelial Cells Involves Multiple 3-O-Sulfotransferase isoforms and a Limiting Precursor Pool". The Journal of Biological Chemistry. 280 (45): 38059–38070. doi:10.1074/jbc.m507997200.
- ↑ Ori, A.; et al. (2008). "The heparanome and regulation of cell function: structures, functions and challenges". Frontiers Biosci. 13: 4309–4338. doi:10.2741/3007. PMID 18508513.
- ↑ Sadir, R.; et al. (1998). "The heparan sulphate binding sequence of interferon-γ increased the on rate of the interferon-γ / interferon-γ receptor complex formation". J. Biol. Chem. 273 (18): 10919–10925. doi:10.1074/jbc.273.18.10919. PMID 9556569.
- ↑ Tong et al, Stimulated neovascularization, inflammation resolution and collagen maturation in healing rat cutaneous wounds by a heparan sulfate glycosaminoglycan mimetic, OTR4120. Wound Repair Regen. 2009 Nov-Dec;17(6):840-52.
- ↑ Tong et al, Heparan sulfate glycosaminoglycan mimetic improves pressure ulcer healing in a rat model of cutaneous ischemia-reperfusion injury. Wound Repair Regen. 2011 Jul;19(4):505-14.
- ↑ Van Neck et al, Heparan sulfate proteoglycan mimetics thrive tissue regeneration: an overview. In Intech book under the working title "Tissue Regeneration", ISBN 978-953-307-876-2 is scheduled for on line publication on Nov 26, 2011”