GlmS glucosamine-6-phosphate activated ribozyme
| glmS glucosamine-6-phosphate activated ribozyme | |
|---|---|
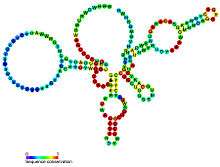 | |
| Predicted secondary structure and sequence conservation of glmS | |
| Identifiers | |
| Symbol | glmS |
| Rfam | RF00234 |
| Other data | |
| RNA type | Cis-reg; riboswitch |
| Domain(s) | Bacteria |
| SO | 0000035 |

The Glucosamine-6-phosphate activated ribozyme ( glmS ribozyme) is an RNA structure that is both a ribozyme, since it catalyzes a chemical reaction, and a riboswitch, since it regulates genes in response to concentrations of a metabolite.[3] It was originally identified using bioinformatics in the 5' untranslated regions of glmS genes.[4] The GlmS enzyme catalyzes the production of glucosamine-6-phosphate (GlcN6P), and the glmS ribozyme is dependent on GlcN6P to achieve catalysis of its own cleavage.[3] This cleavage leads to the degradation of the mRNA that contains the ribozyme, and ultimately lowers production of the GlmS enzyme, encoded by the glmS gene.[5] The GlmS enzyme catalyzes the reaction to synthesize GlcN6P, so it is not required when GlcN6P levels are already high. When GlcN6P levels are low, the ribozyme does not cleave, allowing production of the GlmS enzyme, and ultimately more GlcN6P. In fact, GlcN6P is a cofactor for the cleavage reaction, as it directly participates in the chemical reaction as an acid-base catalyst.[6]
Structure
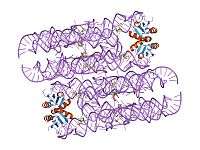
The structure of the GlmS ribozyme has been determined by X-ray crystallography.[1][2] The RNA adopts a double pseudoknotted structure. The cofactor is bound in a solvent-accessible pocket and the structure suggests that the amine group of GlcN6P is involved in the catalytic process.[7][8]
See also
References
- 1 2 Cochrane JC, Lipchock SV, Strobel SA (2007). "Structural investigation of the GlmS ribozyme bound to its catalytic cofactor". Chem. Biol. 14 (1): 97–105. doi:10.1016/j.chembiol.2006.12.005. PMC 1847778
 . PMID 17196404.
. PMID 17196404. - 1 2 Klein DJ, Ferré-D'Amaré AR (2006). "Structural basis of glmS ribozyme activation by glucosamine-6-phosphate". Science. 313 (5794): 1752–6. doi:10.1126/science.1129666. PMID 16990543.
- 1 2 Winkler, WC; Nahvi A; Roth A; Collins JA; Breaker RR (2004). "Control of gene expression by a natural metabolite-responsive ribozyme". Nature. 428 (6980): 281–286. doi:10.1038/nature02362. PMID 15029187.
- ↑ Barrick JE, Corbino KA, Winkler WC, et al. (April 2004). "New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control". Proc. Natl. Acad. Sci. U.S.A. 101 (17): 6421–6. doi:10.1073/pnas.0308014101. PMC 404060
 . PMID 15096624.
. PMID 15096624. - ↑ Collins, JA; Irnov I; Baker S; Winkler WC (2007). "Mechanism of mRNA destabilization by the glmS ribozyme". Genes Dev. 21 (24): 3356–3368. doi:10.1101/gad.1605307. PMC 2113035
 . PMID 18079181.
. PMID 18079181. - ↑ Viladoms, J. L.; Fedor, M. J. (2012). "TheglmSRibozyme Cofactor is a General Acid–Base Catalyst". Journal of the American Chemical Society. 134 (46): 19043–19049. doi:10.1021/ja307021f. PMC 3504194
 . PMID 23113700.
. PMID 23113700. - ↑ Jansen JA, McCarthy TJ, Soukup GA, Soukup JK (2006). "Backbone and nucleobase contacts to glucosamine-6-phosphate in the glmS ribozyme". Nat Struct Mol Biol. 13 (6): 517–23. doi:10.1038/nsmb1094. PMID 16699515.
- ↑ Hampel KJ, Tinsley MM (2006). "Evidence for preorganization of the glmS ribozyme ligand binding pocket". Biochemistry. 45 (25): 7861–71. doi:10.1021/bi060337z. PMID 16784238.