Genome diversity and karyotype evolution of mammals
The 2000s witnessed an explosion of genome sequencing and mapping in evolutionarily diverse species. While full genome sequencing of mammals is rapidly progressing, the ability to assemble and align orthologous whole chromosomal regions from more than a few species is not yet possible. The intense focus on the building of comparative maps for domestic (dogs and cats), laboratory (mice and rats) and agricultural (cattle) animals has traditionally been used to understand the underlying basis of disease-related and healthy phenotypes.
These maps also provide an unprecedented opportunity to use multispecies analysis as a tool to infer karyotype evolution. Comparative chromosome painting and related techniques are very powerful approaches in comparative genome studies. Homologies can be identified with high accuracy using molecularly defined DNA probes for fluorescence in situ hybridization (FISH) on chromosomes of different species. Chromosome painting data are now available for members of nearly all mammalian orders.
It was found that in most orders, there are species with rates of chromosome evolution that can be considered as 'default' rates. It needs to be noted that the number of rearrangements that have become fixed in evolutionary history seems relatively low, due to 180 million years of the mammalian radiation. Thus a record of the history of karyotype changes that have occurred during evolution have been attained through comparative chromosome maps.
Mammalian phylogenomics
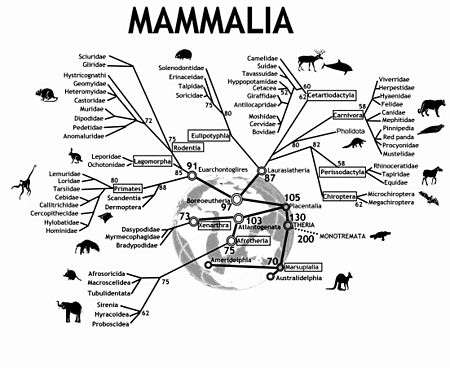
Modern mammals (class Mammalia) are divided into Monotremes, Marsupials, and Placentals. The subclass Prototheria (Monotremes) comprises the five species of egg-laying mammals: platypus and four echidna species. The infraclasses Metatheria (Marsupials) and Eutheria (Placentals) together form the subclass Theria.[4]
In the 2000s understanding of the relationships among eutherian mammals has experienced a virtual revolution. Molecular phylogenomics, new fossil finds and innovative morphological interpretations now group the more than 4600 extant species of eutherians into four major super-ordinal clades: Euarchontoglires (including Primates, Dermoptera, Scandentia, Rodentia, and Lagomorpha), Laurasiatheria (Cetartiodactyla, Perissodactyla, Carnivora, Chiroptera, Pholidota, and Eulipotyphla), Xenarthra, and Afrotheria (Proboscidea, Sirenia, Hyracoidea, Afrosoricida, Tubulidentata, and Macroscelidea).[4] This tree is very useful in unifying the parts of a puzzle in comparative mammalian cytogenetics.
Karyotypes: a global view of the genome
Each gene maps to the same chromosome in every cell. Linkage is determined by the presence of two or more loci on the same chromosome. The entire chromosomal set of a species is known as a karyotype.
A seemingly logical consequence of descent from common ancestors is that more closely related species should have more chromosomes in common. However, it is now widely thought that species may have phenetically similar karyotypes due to genomic conservation. Therefore, in comparative cytogenetics, phylogenetic relationships should be determined on the basis of the polarity of chromosomal differences (derived traits).
Historical development of comparative cytogenetics
Mammalian comparative cytogenetics, an indispensable part of phylogenomics, has evolved in a series of steps from pure description to the more heuristic science of the genomic era. Technical advances have marked the various developmental steps of cytogenetics.
Classical phase of cytogenetics

The first step of the Human Genome Project took place when Tjio and Levan, in 1956, reported the accurate diploid number of human chromosomes as 2n = 46.[6]
During this phase, data on the karyotypes of hundreds of mammalian species (including information on diploid numbers, relative length and morphology of chromosomes, presence of B chromosomes) were described. Diploid numbers (2n) were found to vary from 2n = 6–7 in the Indian muntjac[7] to over 100 in some rodents.[8]
Chromosome banding
The second step derived from the invention of C-, G-, R- and other banding techniques and was marked by the Paris Conference (1971), which led to a standard nomenclature to recognize and classify each human chromosome.[9]
G- and R- banding
The most widely used banding methods are G-banding (Giemsa-banding) and R-banding (reverse-banding). These techniques produce a characteristic pattern of contrasting dark and light transverse bands on the chromosomes. Banding makes it possible to identify homologous chromosomes and construct chromosomal nomenclatures for many species. Banding of homologous chromosomes allows chromosome segments and rearrangements to be identified. The banded karyotypes of 850 mammalian species were summarized in the Atlas of Mammalian Chromosomes.[10]
C-banding and heterochromatin

Karyotype variability in mammals is mainly due to the varying amount of heterochromatin in each mammal. Once the amount of heterochomatin is subtracted from total genome content, all mammals have very similar genome sizes.
Mammalian species differ considerably in heterochromatin content and location. Heterochromatin is most often detected using C-banding.[12] Early studies using C-banding showed that differences in the fundamental number (i.e., the number of chromosome arms) could be entirely due to the addition of heterochromatic chromosome arms. Heterochromatin consists of different types of repetitive DNA, not all seen with C-banding that can vary greatly between karyotypes of even closely related species. The differences of the amount of heterochromatin among congeneric rodent species may reach 33% of nuclear DNA in Dipodomys species,[13] 36% in Peromyscus species,[14] 42% in Ammospermophilus[15] and 60% in Thomomys species where C-value (haploid DNA content) ranges between 2.1 and 5.6 pg.[16][17]
The red viscacha rat (Tympanoctomys barrerae) has a record C-value among mammals—9.2 pg.[18] Although tetrapoidy was first proposed to be a reason for its high genome size and diploid chromosome number, Svartman et al.[19] showed that the high genome size was due to the enormous amplification of heterochromatin. Although one single copy gene was found to be duplicated in its genome,[20] data on absence of large genome segment duplications (single paints of most Octodon degu probes) and repetitive DNA hybridization evidence rules against tetraploidy. The study of heterochromatin composition, repeated DNA amount and its distribution on chromosomes of octodontids is absolutely necessary to define exactly what heterochromatin fraction is responsible for the large genomes of the red viscacha rat.[21]
In comparative cytogenetics, chromosome homology between species was proposed on the basis of similarities in banding patterns. Closely related species often had very similar banding pattern and after 40 years of comparing bands it seems safe to generalize that karyotype divergence in most taxonomic groups follows their phylogenetic relationship, despite notable exceptions.[10][22]
The conservation of large chromosomal segments makes comparison between species worthwhile. Chromosome banding has been a reliable indicator of chromosome homology overall, i.e. that the chromosome identified on the basis of banding actually carries the same genes. This relationship may fail for phylogenetically distant species or species that have experienced extremely rapid chromosome evolution. Banding is still morphological and is not always a foolproof indicator of DNA content.[21]
Comparative molecular cytogenetics
The third step occurred when molecular techniques were incorporated into cytogenetics. These techniques use DNA probes of diverse sizes to compare chromosomes at the DNA level. Homology can be confidently compared even between phylogenetically distant species or highly rearranged species (e.g., gibbons). Using cladistic analysis rearrangements that have diversified the mammalian karyotype are more precisely mapped and placed in a phylogenomic perspective. "Comparative chromosomics" defines the field of cytogenetics dealing with molecular approaches,[30] although "chromosomics" was originally introduced to define the research of chromatin dynamics and morphological changes in interphase chromosome structures.[31]
Chromosome painting or Zoo-FISH was the first technique to have a wide-ranging impact.[32][33][34][35][36] With this method the homology of chromosome regions between different species are identified by hybridizing DNA probes of an individual, whole chromosomes of one species to metaphase chromosomes of another species. Comparative chromosome painting allows a rapid and efficient comparison of many species and the distribution of homologous regions makes it possible to track the translocation of chromosomal evolution. When many species covering different mammalian orders are compared, this analysis can provide information on trends and rates of chromosomal evolution in different branches.
However, homology is only detected qualitatively, and resolution is limited by the size of visualized regions. Thus, the method does not detect all minuscule homologous regions from multiple rearrangements (as between mouse and human). The method also fails to report internal inversions within large segments. Another limitation is that painting across great phylogenetic distance often results in a decreased efficiency. Nevertheless, the use of painting probes derived from different species combined with comparative sequencing projects help to increase the resolution of the method.[21]
In addition to sorting, microdissection of chromosomes and chromosome regions was also used to obtain probes for chromosome painting. Best results were obtained when a series of microdissection probes covering the total human genome were localized on anthropoid primate chromosomes via multicolor banding (MCB).[37][38] However a limitation of MCB is that it can only be used within a group of closely related species ("phylogenetic" resolution is too low). Spectral karyotyping (SKY) and MFISH—the ratio labeling and simultaneous hybridization of a complete chromosomal set have similar drawbacks and little application outside of clinical studies.[21]
Comparative genomics data including chromosome painting confirmed the substantial conservation of mammalian chromosomes.[36] Total human chromosomes or their arms can efficiently paint extended chromosome regions in many placentals down to Afrotheria and Xenarthra. Gene localization data on human chromosomes can be extrapolated to the homologous chromosome regions of other species with high reliability. Usefully, humans express conserved syntenic chromosome organization similar to the ancestral condition of all placental mammals.
Post-genomic time and comparative chromosomics
After the Human Genome Project researchers focused on evolutionary comparisons of the genome structures of different species. The whole genome of any species can be sequenced completely and repeatedly to obtain a comprehensive single-nucleotide map. This method makes it possible to compare genomes for any two species regardless of their taxonomic distance.
Sequencing efforts provided a variety of products useful in molecular cytogenetics. Fluorescence in situ hybridization (FISH) with DNA clones (BAC and YAC clones, cosmids) allowed the construction of chromosome maps at a resolution of several megabases that could detect relatively small chromosome rearrangements. A resolution of several kilobases can be achieved on interphase chromatin. A limitation is that hybridization efficiencies decrease with increasing phylogenetic distance.
Radiation hybrid (RH) genome mapping is another efficient approach. This method includes the irradiation of cells to disrupt the genome into the desired number of fragments that are subsequently fused with Chinese hamster cells. The resulting somatic cell hybrids contain individual fragments of the relevant genome. Then, 90–100 (sometimes, more) clones covering the total genome are selected, and the sequences of interest are localized on the cloned fragments via the polymerase chain reaction (PCR) or direct DNA–DNA hybridization. To compare the genomes and chromosomes of two species, RHs should be obtained for both species.[21]
Sex chromosome evolution
In contrast to many other taxa, therian mammals and birds are characterized by highly conserved systems of genetic sex determination that lead to special chromosomes, i.e. the sex chromosomes. Although the XX/XY sex chromosome system is the most common among eutherian species, it is not universal. In some species X-autosomal translocations result in the appearance of "additional Y" chromosomes (for example, XX/XY1Y2Y3 systems in black muntjac).[39][40]
In other species Y-autosomal translocations lead to appearance of additional X chromosomes (for example, in some New World primates such as howler monkeys). Regarding this aspect, rodents again represent a peculiar derived group, comprising the record number of species with non-classical sex chromosomes such as the wood lemming, the collared lemming, the creep vole, the spinous country rat, the Akodon and the bandicoot rat.[41]
References
 This article incorporates text from a scholarly publication published under a copyright license that allows anyone to reuse, revise, remix and redistribute the materials in any form for any purpose: Graphodatsky, A. S.; Trifonov, V. A.; Stanyon, R. (2011). "The genome diversity and karyotype evolution of mammals". Molecular Cytogenetics. 4: 22. doi:10.1186/1755-8166-4-22. PMC 3204295
This article incorporates text from a scholarly publication published under a copyright license that allows anyone to reuse, revise, remix and redistribute the materials in any form for any purpose: Graphodatsky, A. S.; Trifonov, V. A.; Stanyon, R. (2011). "The genome diversity and karyotype evolution of mammals". Molecular Cytogenetics. 4: 22. doi:10.1186/1755-8166-4-22. PMC 3204295 . PMID 21992653. Please check the source for the exact licensing terms.
. PMID 21992653. Please check the source for the exact licensing terms.
- ↑ This tree depicts historic divergence relationships among the living orders of mammals. The phylogenetic hierarchy is a consensus view of several decades of molecular genetic, morphological and fossil inference (see for example,[2][3]). Double rings indicate mammalian supertaxa, numbers indicate the possible time of divergences.
- ↑ Murphy, W. J.; Pringle, T. H.; Crider, T. A.; Springer, M. S.; Miller, W. (2007). "Using genomic data to unravel the root of the placental mammal phylogeny". Genome Research. 17: 413–421. doi:10.1101/gr.5918807. PMC 1832088
 . PMID 17322288.
. PMID 17322288. - ↑ Bininda-Emonds, O. R.; Cardillo, M.; Jones, K. E.; MacPhee, R. D.; Beck, R. M.; Grenyer, R.; Price, S. A.; Vos, R. A.; Gittleman, J. L.; Purvis, A. (2007). "The delayed rise of present-day mammals". Nature. 446: 507–512. doi:10.1038/nature05634. PMID 17392779.
- 1 2 Murphy, W. J.; Eizirik, E.; Johnson, W. E.; Zhang, Y. P.; Ryder, O. A.; O'Brien, S. J. (2001). "Molecular phylogenetics and the origins of placental mammals". Nature. 409: 614–618. doi:10.1038/35054550. PMID 11214319.
- ↑ a. Metaphase spread of the Indian muntjac (Muntiacus muntjak vaginalis, 2n = 6, 7), the species with the lowest chromosome number. b. Metaphase spread of the Viscacha rat (Tympanoctomys barrerae, 2n = 102), the species with the highest chromosomal number. c. Metaphase spread of the Siberian roe deer (Capreolus pygargus, 2n = 70 + 1-14 B's), the species with additional, or B- chromosomes. d. Metaphase spread of the Transcaucasian Mole Vole female (Ellobius lutescens, 2n = 17, X0 in both sexes).
- ↑ Tjio, HJ LA; Levan, Albert (1956). "The chromosome numbers of man". Hereditas. 42: 1–6. doi:10.1111/j.1601-5223.1956.tb03010.x.
- ↑ Wurster, D. H.; Benirschke, K. (1970). "Indian muntjac, Muntiacus muntjak: a deer with a low diploid chromosome number". Science. 168 (3937): 1364–1366. doi:10.1126/science.168.3937.1364. PMID 5444269.
- ↑ Contreras, L. C.; Torresmura, J. C.; Spotorno, A. E. (1990). "The Largest Known Chromosome-Number for a Mammal, in a South-American Desert Rodent". Experientia. 46 (5): 506–508. doi:10.1007/BF01954248. PMID 2347403.
- ↑ "Paris Conference (1971): Standardization in human cytogenetics". Cytogenetics. 11 (5): 317–362. 1972. doi:10.1159/000130202. PMID 4647417.
- 1 2 S. J. O'Brien, W. G. N.; Menninger, J. C. (2006). "Atlas of Mammalian Chromosomes".
- ↑ a. C-banded chromosomes of the Eurasian shrew (Sorex araneus, 2n = 21), example of the smallest amount of heterochromatic bands in mammalian genome. b. C-banded chromosomes of the ground squirrel (Spermophilus erythrogenys, 2n = 36) with very large centomeric C-bands. c. C-banded chromosomes of the marbled polecat (Vormela peregusna, 2n = 38) with the largest additional heterochromatic arms on some autosomes. d. C-banded chromosomes of the Amur hedgehog (Erinaceus amurensis, 2n = 48) with the very large telomeric C-bands on autosomes. e. C-banded chromosomes of the Eversmann's hamster (Allocricetulus eversmanni, 2n = 26) with pericentomeric C-bands on the X and Y chromosomes. f. C-banded chromosomes of the southern vole (Microtus rossiaemeridionalis, 2n = 54) with very large C-bands on both sex chromosomes.
- ↑ Hsu, T. C.; Arrighi, F. E. (1971). "Distribution of constitutive heterochromatin in mammalian chromosomes". Chromosoma. 34 (3): 243–253. doi:10.1007/BF00286150. PMID 5112131.
- ↑ Hatch, F. T.; Bodner, A. J.; Mazrimas, J. A.; Moore, D. H. (1976). "Satellite DNA and cytogenetic evolution. DNA quantity, satellite DNA and karyotypic variations in kangaroo rats (genus Dipodomys)". Chromosoma. 58 (2): 155–168. doi:10.1007/BF00701356. PMID 1001153.
- ↑ Deaven, L. L.; Vidal-Rioja, L.; Jett, J. H.; Hsu, T. C. (1977). "Chromosomes of Peromyscus (rodentia, cricetidae). VI. The genomic size". Cytogenet Cell Genet. 19 (5): 241–249. doi:10.1159/000130816. PMID 606503.
- ↑ Mascarello, JT MJ; Mazrimas, J. A. (1977). "Chromosomes of antelope squirrels (genus Ammospermophilus): a systematic banding analysis of four species with unusual constitutive heterochromatin". Chromosoma. 64 (3): 207–217. doi:10.1007/BF00328078.
- ↑ Patton, J. L.; Sherwood, S. W. (1982). "Genome evolution in pocket gophers (genus Thomomys). I. Heterochromatin variation and speciation potential". Chromosoma. 85 (2): 149–162. doi:10.1007/BF00294962. PMID 7117026.
- ↑ Sherwood, S. W.; Patton, J. L. (1982). "Genome evolution in pocket gophers (genus Thomomys). II. Variation in cellular DNA content". Chromosoma. 85 (2): 163–179. doi:10.1007/BF00294963. PMID 7117027.
- ↑ Gallardo, M. H.; Bickham, J. W.; Honeycutt, R. L.; Ojeda, R. A.; Kohler, N. (1999). "Discovery of tetraploidy in a mammal". Nature. 401 (6751): 341. doi:10.1038/43815.
- ↑ Svartman, M.; Stone, G.; Stanyon, R. (2005). "Molecular cytogenetics discards polyploidy in mammals". Genomics. 85 (4): 425–430. doi:10.1016/j.ygeno.2004.12.004. PMID 15780745.
- ↑ Gallardo, M. H.; Gonzalez, C. A.; Cebrian, I. (2006). "Molecular cytogenetics and allotetraploidy in the red vizcacha rat, Tympanoctomys barrerae (Rodentia, Octodontidae)". Genomics. 88 (2): 214–221. doi:10.1016/j.ygeno.2006.02.010. PMID 16580173.
- 1 2 3 4 5 Graphodatsky, A. S.; Trifonov, V. A.; Stanyon, R. (2011). "The genome diversity and karyotype evolution of mammals". Molecular Cytogenetics. 4: 22. doi:10.1186/1755-8166-4-22. PMC 3204295
 . PMID 21992653.
. PMID 21992653. - ↑ Graphodatsky, A. S. (2006). "Conserved and variable elements of mammalian chromosomes". Cytogenetics of animals: 95–124.
- ↑ a. Reconstructed karyotype of the ancestral Eutherian genome.[24] Each chromosome is assigned a specific color. These colors are used for mark homologies in idiograms of chromosomes of other species (5b–5i) b. Idiogram of chicken (Gallus gallus domesticus, 2n = 78) chromosomes. The reconstruction is based on alignments of chicken and human genome sequences.[25] c. Idiogram of short-tailed opossum (Monodelphis domestica, 2n = 18) chromosomes. The reconstruction is based on alignments of opossum and human genome sequences.[25] d. Idiogram of aardvark (Orycteropus afer, 2n = 20) chromosomes. The reconstruction is based on painting data.[24] e. Idiogram of mink (Mustela vison, 2n = 30) chromosomes. The reconstruction is based on painting data.[26][27] f. Idiogram of the Red fox (Vulpes vulpes, 2n = 34 + 0-8 B's) chromosomes. The reconstruction is based on painting and mapping data.[28][29] g. Reconstructed karyotype of the ancestral Sciuridae (Rodentia) genome, based on painting data (Li et al., 2004). h. Idiogram of the House mouse (Mus musculus, 2n = 40) chromosomes. The reconstruction is based on alignments of Mus and human genome sequences.[25] i. Idiogram of human (Homo sapiens, 2n = 46) chromosomes.
- 1 2 Stanyon, R.; G. Stone; M. Garcia; L. Froenicke (2003). "Reciprocal chromosome painting shows that squirrels, unlike murid rodents, have a highly conserved genome organization". Genomics. 82 (2): 245–249. doi:10.1016/S0888-7543(03)00109-5. PMID 12837274.
- 1 2 3 "Ensembl Genome Browser".
- ↑ Hameister, H.; Klett, C.; Bruch, J.; Dixkens, C.; Vogel, W.; Christensen, K. (1997). "Zoo-FISH analysis: the American mink (Mustela vison) closely resembles the cat karyotype". Chromosome Res. 5 (1): 5–11. doi:10.1023/A:1018433200553. PMID 9088638.
- ↑ Graphodatsky, A. S.; Yang, F.; Perelman, P. L.; O'Brien, P. C. M.; Serdukova, N. A.; Milne, B. S.; Biltueva, L. S.; Fu, B.; Vorobieva, N. V.; Kawada, S. I. (2002). "Comparative molecular cytogenetic studies in the order Carnivora: mapping chromosomal rearrangements onto the phylogenetic tree". Cytogenetic and Genome Research. 96 (1–4): 137–145. doi:10.1159/000063032. PMID 12438790.
- ↑ Yang, F.; O'Brien, P. C.; Milne, B. S.; Graphodatsky, A. S.; Solanky, N.; Trifonov, V.; Rens, W.; Sargan, D.; Ferguson-Smith, M. A. (1999). "A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps". Genomics. 62 (2): 189–202. doi:10.1006/geno.1999.5989. PMID 10610712.
- ↑ Graphodatsky, A. S.; Kukekova, A. V.; Yudkin, D. V.; Trifonov, V. A.; Vorobieva, N. V.; Beklemisheva, V. R.; Perelman, P. L.; Graphodatskaya, D. A.; Trut, L. N.; Yang, F. T. (2005). "The proto-oncogene C-KIT maps to canid B-chromosomes". Chromosome Research. 13 (2): 113–122. doi:10.1007/s10577-005-7474-9. PMID 15861301.
- ↑ Grafodatskii, A. S. (2007). "[Comparative chromosomics]". Mol Biol (Mosk). 41: 408–422.
- ↑ Claussen, U. (2005). "Chromosomics". Cytogenet Genome Res. 111 (2): 101–106. doi:10.1159/000086377. PMID 16103649.
- ↑ Wienberg, J.; Jauch, A.; Stanyon, R.; Cremer, T. (1990). "Molecular cytotaxonomy of primates by chromosomal in situ suppression hybridization". Genomics. 8 (2): 347–350. doi:10.1016/0888-7543(90)90292-3. PMID 2249853.
- ↑ Telenius, H.; Pelmear, A. H.; Tunnacliffe, A.; Carter, N. P.; Behmel, A.; Fergusonsmith, M. A.; Nordenskjold, M.; Pfragner, R.; Ponder, B. A. J. (1992). "Cytogenetic Analysis by Chromosome Painting Using Dop-Pcr Amplified Flow-Sorted Chromosomes". Gene Chromosome Canc. 4 (3): 257–263. doi:10.1002/gcc.2870040311.
- ↑ Scherthan, H.; Cremer, T.; Arnason, U.; Weier, H. U.; Limadefaria, A.; Fronicke, L. (1994). "Comparative Chromosome Painting Discloses Homologous Segments in Distantly Related Mammals". Nature Genetics. 6 (4): 342–347. doi:10.1038/ng0494-342. PMID 8054973.
- ↑ Ferguson-Smith, M. A. (1997). "Genetic analysis by chromosome sorting and painting: phylogenetic and diagnostic applications". Eur J Hum Genet. 5 (5): 253–265. PMID 9412781.
- 1 2 Ferguson-Smith, M. A.; V. Trifonov (2007). "Mammalian karyotype evolution". Nat Rev Genet. 8 (12): 950–962. doi:10.1038/nrg2199. PMID 18007651.
- ↑ Mrasek, K.; Heller, A.; Rubtsov, N.; Trifonov, V.; Starke, H.; Rocchi, M.; Claussen, U.; Liehr, T. (2001). "Reconstruction of the female Gorilla gorilla karyotype using 25-color FISH and multicolor banding (MCB)". Cytogenetics and Cell Genetics. 93 (3–4): 242–248. doi:10.1159/000056991. PMID 11528119.
- ↑ Mrasek, K.; Heller, A.; Rubtsov, N.; Trifonov, V.; Starke, H.; Claussen, U.; Liehr, T. (2003). "Detailed Hylobates lar karyotype defined by 25-color FISH and multicolor banding". International Journal of Molecular Medicine. 12 (2): 139–146. doi:10.3892/ijmm.12.2.139. PMID 12851708.
- ↑ Yang, F.; Carter, N. P.; Shi, L.; Ferguson-Smith, M. A. (1995). "A comparative study of karyotypes of muntjacs by chromosome painting". Chromosoma. 103 (9): 642–652. doi:10.1007/BF00357691. PMID 7587587.
- ↑ Huang, L.; Chi, J.; Wang, J.; Nie, W.; Su, W.; Yang, F. (2006). "High-density comparative BAC mapping in the black muntjac (Muntiacus crinifrons): molecular cytogenetic dissection of the origin of MCR 1p+4 in the X1X2Y1Y2Y3 sex chromosome system". Genomics. 87 (5): 608–615. doi:10.1016/j.ygeno.2005.12.008. PMID 16443346.
- ↑ Fredga, K. (1983). "Aberrant sex chromosome mechanisms in mammals. Evolutionary aspects". Differentiation. 23 Suppl: S23–30. doi:10.1007/978-3-642-69150-8_4. PMID 6444170.
