Genetic studies on Bulgarians
The Bulgarians are part of the Slavic ethnolinguistic group as a result of migrations of Slavic tribes to the region since the 6th century AD and the subsequent linguistic assimilation of other populations.[1][2][3][4]
Genetic admixture analyses based on data of several individuals of modern populations, estimate that the genes of the Bulgarians have two sources and a main admixture event between 1000 and 1600 YBP. The genes that are provisionally or indicatively to be associated with the Balto-Slavic donor group constitute up to over 40% of the total,[5] i.e. Belarusian-like, of which a Lithuanian-like admixture is estimated at 23.2% and a Polish-like admixture at 19.3%.[6] In DNA.Land, an ancestral category named "North Slavic" ancestry(which includes Baltic) comprises an average of 25-30% in Bulgarians. Early gene flows between southeastern and northeastern Europe make it difficult to obtain a correct estimate, although young enough identical by descent segments confirmed such connection and that the East and West Slavs share more identical by descent segments with South Slavs than with Greeks, inter-Slavic populations(a group of Romanians, Gagauz and Hungarians) and Balts.[7] Genetically and linguistically the Slavic ancestors of the modern South Slavs had divided mainly into two groups, each of which took a migration stream either through the west or the east of the Carpathian Mountains.[8] The western Balkans was settled with Sclaveni, the east with Antes.[9] Haplogroup R1a, the major haplogroup among Slavic tribes, reveals that the haplogroup of the Serbo-Croat group is mainly constituted by R1a-L1280 or R1a-CTS3402, while the Bulgaro-Macedonian is exclusively made up of the R1a-L1029.[10] Linguistic evidence backs similar division of Western and Eastern South Slavic, further showing that the Eastern South Slavic group demonstrates closer affinity and some irreplaceable key features present only in the Lechitic dialects.[11]
The modern Bulgarians and all other South Slavs(excl. Slovenians) are characterized by a prevailing genetic substrate that is different than that of the North Slavs.[7] The two groups are today separated, sharing a modest gene flow.[7][12] This phenomena of two genetically distinct groups of Slavic peoples is explained by the assimilation of more numerous previous indigenous populations by the medieval Slavic settlers in the Balkans.[7][12] Over 50% from Bulgarian genetic legacy is Mediterranean, about the half of which resembles the Caucasian, Middle Eastern and to a lesser extent the North African genetics.[5]
Around 4% of Bulgarian genes are derived outside of Europe and the Middle East or are of undetermined origin (by 858 CE), of which 2.3% are from Northeast Asia and correspond to Asian tribes such as Bulgars,[13] a consistent very low frequency for Eastern Europe as far as Uralic-speaking Hungarians. The percentage, however, may vary by regions as the less numerous Bulgars were concentrated around the northeast failing to settle throughout the country numerous tribes as the significant donor tribes.
Y DNA
Bulgarians, as some of their neighbours show the highest diversity of haplogroups in Europe, marked by significant (> 10%) frequencies of 5 major haplogroups (compared to Atlantic Europe, dominated by > 50% R1b). Most Bulgarians belong to three unrelated haplogroups, 20% of whom to I-M423 (I2a1b), 18% to E-V13 (E1b1b1a1b1a) and 18% to R-M17 (R1a1a), but the biggest part belongs to macro-haplgoroup R (~28%). The major haplogroups, groupped by age of around 20 kya, are:[14]
- Haplogroup I-L460 (I2a) is presented at levels 21.9% [15] according to 808 Bulgarian male samples of the largest-scale study from 2013. By higher levels are defined the profiles of Ukrainians and all South Slavs other than Slovenians.[16] Evidence points to European origin for macro-haplogroup I, and Levantine for its immediate ancestor- IJ. Its exclusive and now patchy distribution within Europe suggested a very early entry in to Europe during Palaeolithic colonization, which was confirmed by the lack of its ancient DNA outside of the continent and ~13,000 years old European Cro-Magnon remains belonging to I2a.[17] I2a2 is the most frequent haplogroup of European male remains dated to the Metal Ages, while I2a1 and I2a1b are most common on Mesolithic remains,[17][18] as such they were the primary haplogroups of pre-historic European hunter-gatherers. Initially a Holocene expansion of I2a in Southeastern Europe is supposed;[19] however Dinaric is descended by several 'only child' sublclades and it is suggested that its most recent common ancestor is aged only 2200 years[20] making it the youngest and most common micro-group.
- I2a is recorded to be the dominant haplogroup in the former Sofia-city, Sofia, Plovdiv and Varna province between 23-33%, dropping under 10% in central Bulgaria. Bulgarian Hg I2a most often belongs specifically to the P37.2, M423 branch ("Hg I2a1b"), representing 20% of Bulgarian males.[15] According to some data, Bulgarian males who belong to M423, belong to CTS10228 Dinaric (I2a1b2a1), the prevalent clade in eastern Europe, and none of them belongs to the Disles L161 branch or the CTS595 branch found in west Europe and Sardinia.[21] It makes up an absolute majority among the highest populations. Many authors claim that the higher ratio of I2a1b over R1a that is observed in the Balkans today is the founder effect and was present before the Slavic expansion, owed to indigenous tribes,[7] and especially for the highest frequency in the Western Balkan area where Dinaric-South haplotypes prevail. However, the largest haplotype group among Bulgarians is Dinaric-North (75%), which is prevalent among all North Slavs and may suggest a recent migration north of the Danube river. Anyway, any evidence for such migration is yet not observed in connection to modern populations as more than 90% of the traced Dinaric-North Bulgarian haplotypes belong to the SNP group Z17855, being the prevailing clade in Bulgarians, somehow significant in the western Balkans and rare elsewhere observed.[21][22] These SNP groups separated before the Slavic expansion in the Balkans. Some authors suggested that I2a-Dinaric is pre-Slavic in the region and owed to the Thracians.[23] According to large datasets the dominant Bulgarian Z17855 SNP is even absent in north Slavic DNA projects. The rest ~2% of Bulgarian males belong to the subclade M223 (I2a2a).
- Haplogroup E-V68 (E1b1b1a) is presented at levels 19.6% per 880 samples.[24] The ultimate origin of E-V68 points to northeastern Africa, specifically near the Nile and Lake Alexandria.[25] Thus this haplogroup represents a more recent Bronze Age "out of Africa" movement into Europe via the Balkans. The macro-haplogroup E still prevails in most of the African continent, but through the long-term migrations the sub-Saharian maternal lineage Hg L was lost lacking completely in the Balkans. Holocene movement into the Near East is proposed, then several thousand years ago, a movement into the Balkans.[25] All V68-positive Bulgarians belong to its M78 subclade, which is the prevailing haplogroup in most of northeast Africa and around the Balkans. The presently mostly European V13 (E1b1b1a1b1a) originated in western Asia according to the most plausible scenario[26] and is presented at ~18% among Bulgarian males.[15] According to deeply traced data its internal structure is divided among Z5016, Z5017 and S7461.[21] Recent findings of V13 in a Neolithic context in Iberia (dated to ~ 7 kya) give a terminus ante quem.[27] However, it might have really begun to expand in the Balkans somewhat later, perhaps during the population growth of the Bronze Age.[28] It resulted to be the dominant haplogroup in the former Burgas, Lovech, Montana and Razgrad province, from 20% to 38%. Like I-P37 above, it is rather limited to Europe but peaks in the Balkans, only for the Albanians, Greeks, Macedonians, various Romani, Montenegrins, Serbs and Romanians are recorded higher levels than Bulgarians.[29] It has been detected on ancient Thracian remain from Bulgaria. An odd low frequency of Haplogroup E of 10% is recorded in the capital Sofia, which is the lowest on the Balkan peninsula after Croatia and the same level is observed as far as Berlin.[30]
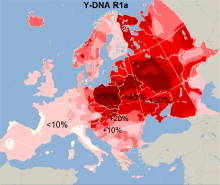
- Haplogroup R-M420 (R1a) is identified at 17.6%[15] per 808 samples. It is the dominant group among the North Slavs, Slovenians, Hungarians and the most common haplogroup in Asia. The overall evidence suggests that the macro-haplogroup R arose in southern or central Asia descending from Haplogroup IJK. The subsequent path into Europe, and the major settlement is thought to have happened in the Bronze Age by the Kurgan hypothesis, R1a and R1b clades are found at minority levels in Europe since the Mesolithic. It has been revealed that the R1a branch Z282 that is limited to Eastern Europe and separated from their Asian relative ~5000 [20] years ago, makes up 96% of Bulgarian R1a, while the most common branch from China to Anatolia (Z93) makes up the rest 4%.[31] As such, the R1a frequency may only be the result of ultimate descendants of ancient eastern European tribes, namely Balto-Slavs, who are the early Slavs and possibly the Thracians. Divided by the largest branches, per 880 samples the levels of the branches of R1a are - M458 - 7.4%, CTS1211(Z280) - 7.1%, Z92(Z280) - 1.9% and Z93 - only 0.7%.[31] According to 100 samples M458 carriers constitute 56% of Bulgarian R1a carriers.[32] Deeply traced data reveals that 90% of the sampled Bulgarian carriers of the M458 clade are carriers of the L1029 micro-clade (R1a1a1b1a1b1), which is 2-3,000 years old, and the L1029 clade of M458 alone accounts for 50% of all Bulgarian R1a per ~250 samples.[10][21] The Z92 component of the Bulgarians is also much lower than that of the East Slavs and more similar to that of the West. All branches are consistently outnumbered by M458 throughout whole eastern and central Bulgaria.[15] M458 is the dominant R1a clade in the regions roughly corresponding to the area of Bulgarian dialects that share most similarities with Polish dialects, the eastern dialects[15] and the Slavic dialects in Greece. R1a with a prevailing M458 (17%) component makes up the majority in the former Haskovo province with the highest frequency on the Balkan Peninsula (29%), while in the former Montana province it is dominant(23%) with dominant Z280 (19%) component.[15] A drastical drop of the ratio of M458 in the northwest is observed. R1a in no province drops under 10%.
- A fact is that the early habitat of the mentioned Slavic tribes by the ancient authors, that are said to have partially moved to Bulgaria (Antes, Severians and Dregovichs/Drougoubitai), was described as being in present Ukraine and Belarus. Many of the tribes there were Lechites, as were the Slavic tribes that settled in Bulgaria by their linguistic traits, moreover today backed by genetic traits. The occasional prevalence of the M458 clade in other Slavic than western countries is traced to historical Lechites as in the case of Radimichs, Polans and Vyatichi, which were assimilated by later East Slavic tribes. Although Lechitic language's now gone from this area, a genetic offshot still remains in the Chernigov, Polesye and northern Dnieper areas, which today are the only areas in Belarus and Ukraine, where M458 is occasionally the prevailing clade,[7][33] as well its microclade L1029.[10] A solid genetic evidence for that the haplogroup R1a of the Bulgarians is for the most part descended from Slavs rather than Thracians and exactly from the Polesye area, is that the L1029 clade among Bulgarians is carried by half the Bulgarian carriers of R1a, and in the case is entirely constituted of two 2000 years old subclades YP417 and YP263,[21] which in turn make up the same majority in several northeastern Euroepan countries. As among Bulgarians, the YP417 clade constitutes the absolute majority of L1029 clade of the sampled people from Belarus and Ukraine, but in contrast in Poland these are not the most common detected clades of L1029. The clade of M458 in Belarus and Ukraine is mostly L1029, but in Poland it is more often L260.[34][35][36][37]
- Haplogroup R-M343 (R1b): present in Bulgarians at 10.7%.[15] R1b is the most frequently occurring haplogroup around Ural and Chad, in most of western Europe and the adjacent islands. A Balkan entry of R1b into Europe is a major theory. The Bulgarian internal structure is heterogeneous and 4% of Bulgarian males carry western European subclades.[15] 3% are carriers of the 'Italo-Atlantic' Proto-Celtic branch P312, of which 2% of U152. Another 1 percent belongs to the U106 branch that corresponds with the spread of Germanic peoples.[15] The ancestral L23 and Z2103 branch show a clear relationship with Anatolia and the Near East.[38] The branch turned to be the dominant clade of the Yamna culture[39] in far eastern Europe. In addition to the Middle East it is currently the dominant clade of R1b there in parts of central and east Europe.[40] Most Bulgarians (6%) belong to the branch, the majority of them belong to its subclade Z2110[21] (R1b1a1a2a2c1a), which today is likely limited only around Europe. The Bulgarian STR markers are closest to the Romanian.[41]
- Haplogroup J-M172 (J2) is presented at levels 10.5%.[24] Higher levels of it are found as far as Hungarians, Romanians, Bosniaks,[42] Austrians and Italians, while Anatolia and the surroundings are dominated by the group.[43] Whilst its origin is north Levantine, its current pattern reflects more recent events connecting the Aegean and western Anatolia during the Copper and Bronze Ages, as well as Greek and Phoenician colonization around the Mediterranean. Several subclades within J2 are present: J-M410 (J2a) is represented at 6%, Balkan J-M12 (J2b) at 4%[15] up to 11% in Burgas(prevailing). The prevailing is the L26 deep subclade of J2a, it is furtherly divided into M67, M92, L24 and other clades.[21]
- Finally, there are also some other Y-DNA Haplogroups presented at a lower levels among Bulgarians ~ 20% all together, as G-P15 (G2a) at ~5%, I-M253 (I1)at ~4% of which L22, Z58 and Z63, many of Scandinavian origin,[21] J-M267 (J1) at ~3.5%, E-M34 (E1b1b1b2a1) at ~2%, T-M70 (T1a) at ~1.5%, at less than 1% Haplogroup C-M217 (C2), H-M82 (H1a1), N-M231 (N), Q-M242 (Q), L-M61 (L), I-M170 (I*), E-M96 (E*) excl. M35, R-M124 (R2a), E-M81. (E1b1b1b1a), E-M35 (E1b1b1*).[15]
| Population | N= | C2 (%) | E1b1b1a1 (%) | E1b1b1b2a1 (%) | E* (%) | G2a (%) | H1a1 (%) | I1 (%) | I2a (%) | I* (%) |
J1 (%) | J2 (%) | L (%) | N (%) | Q (%) | R1a (%) | R1b (%) | T1a (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M217 | M78 | M34 | M96 | P15 | M82 | M253 | L460 | M170 | M267 | M172 | M61 | M231 | M242 | M420 | M343 | M70 | ||
| Burgas | 45 | 20 | 4 | 2 | 4 | 16 | 6 | 18 | 11 | 18 | ||||||||
| Haskovo | 41 | 29 | 2 | 2 | 7 | 2 | 10 | 29 | 15 | 2 | ||||||||
| Lovech | 62 | 24 | 2 | 5 | 3 | 16 | 3 | 18 | 2 | 2 | 19 | 5 | 2 | |||||
| Montana | 80 | 1 | 24 | 2 | 4 | 6 | 19 | 5 | 8 | 1 | 23 | 6 | 1 | |||||
| Plovdiv | 159 | 21 | 2 | 1 | 4 | 1 | 6 | 23 | 1 | 2 | 9 | 1 | 1 | 16 | 12 | 1 | ||
| Sofia City | 59 | 8 | 2 | 3 | 3 | 3 | 27 | 2 | 7 | 9 | 20 | 14 | 2 | |||||
| Sofia Reg | 257 | 1 | 17 | 2 | 7 | 4 | 25 | 4 | 11 | 18 | 9 | 1 | ||||||
| Razgrad | 21 | 38 | 14 | 10 | 10 | 10 | 14 | 5 | ||||||||||
| Varna | 15 | 20 | 7 | 7 | 33 | 7 | 13 | 13 | ||||||||||
| unknown | 69 | 1 | 15 | 1 | 7 | 1 | 6 | 26 | 7 | 1 | 3 | 12 | 14 | 6 | ||||
| Total | 808 | 20 | 2 | 5 | 1 | 4 | 22 | 3 | 10 | 18 | 11 | 2 |
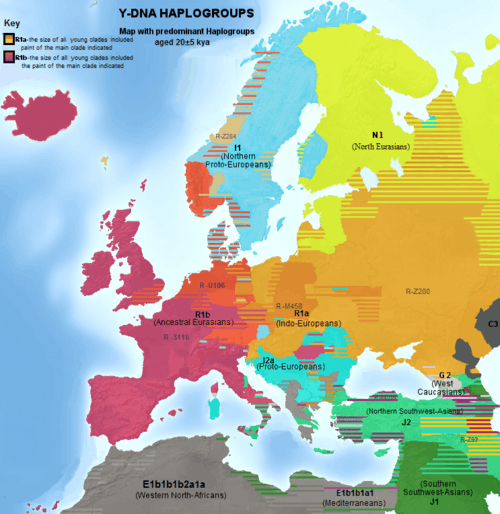
The overall profile of 808 Bulgarian samples, according to a level of phylogenetic analysis calculating distribution of hgs R1a1a7, R1а1, R1b1a2, R2a, I, E1b1, E1b1b1, E1b1b1a, E1b1b1b, J2b, J2a, J2a1b, J1, G, T, NO, C, H, Q, L, A and B, is positioned nearest to the Romanians per 147 their samples,[15] also backed by studies as early as 2000.[44] However the analysis of the Bulgarian study showed inaccuracy in some aspects using population datasets which proved to contain genetic drift according to alternative more extensive studies on these populations. Furthermore, the analysis did not even involve the populations Macedonians, Serbians, Montenegrins and Slovaks. It is unclear why the extensive datasets and the most proximal Slavic populations to the Bulgarians were excluded from the phylogenetic analysis of the Bulgarian study selectively. The study of the 149 Romanians by whose data they came out most proximal to Bulgarians concludes that Romanians are closer to Ukrainians and Hungarians than to the Bulgarian group sampled by the study.[45] Bulgarians are situated nearer to populations such as Berbers and Sudanese than to Turks and Poles, because all subclades of J and E-M78 are counted together.[15] This situation would not be the same if the analysis calculates distance by considering more recent subclades, e.g. J2 and J1 instead of J, or EV13 instead of EM78, similarly Y-DNA analyses of distribution of haplogroups of pre-Mesolithic age determine Norwegians and Sardinians more proximal to Bulgarians than Poles and Russians.[46] Some central Europeans are situated very distant per data of the phylogenetic Bulgarian study because of scarce datasets containing remarkable genetic drift. For most compared European peoples datasets were taken from Battaglia (2008) which does not involve more than 100 samples per population.[47] The largest-scale study of the Hungarians (n=230) determined that[48] the remaining Finno-Urgic peoples are genetically their furthest populations, and clearly confirmed that the closest Europeans to the Hungarians are the Bulgarians, however the same study determines the Yugoslavs as the nearest population to Bulgarians.[49] Similarly, despite linguistic influence, the contribution of Uralics to the modern Hungarian genetic pool is weak as the contribution of Bulgars to the modern Bulgarian genetic pool is hardly recognisable.[50] According to DNA data for 17 Y-chromosomal STR loci in Macedonians, the Macedonian population has the lowest genetic distance against the Bulgarian population (0.0815).[51] Other Y-DNA studies considered the Bulgarians closest either to Macedonians, Serbs, Bulgarian Turks or Gagauzes, followed either by some of these or by Romanians, or Bosniaks.[12][52][53][54]
A phylogenetic analysis determines that the population of Haskovo Province has shorter genetic distance against the population of the Czech Republic than to the Bulgarian provinces, and that only the population of Burgas Province is closer to Haskovo than the Hungarian population, furthermore only datasets of two more Balkan or Slavic foreign populations(Greece and Croatia) are used and all other Slavic populations are excluded from this analysis.[55]
According to an older study of 127 Bulgarian males, frequencies are the following: 30% R (17% R1a, 11% R1b, 2% R*); 27.5% I; 20% E; 18% J; 1.5% G; 1.5% H; 1% T.[56]
According to another study involving 126 Bulgarian males, frequencies are the following: 30% I (25.5% I2a, 4% I1); 20,5% E; 17.5% R (R1b 11%, R1a 6%); 17.5% J (16% J2); 5.5% G; 4% Q; 1% L; 1% T; unknown 3%.[57]
According to another study involving 100 Bulgarian males, frequencies are the following: 34% I (29% I2a, 3% I1); 30% R (16% R1a, 14% R1b); 21% E (20% E1b1b1a); 9% J; 2% G; 2% T; 1% N.[58]
According to 250 Bulgarian samples from FTDNA, frequencies are the following: 27% I (20% I2a1, 4% I1, 2% I2a2, 1% I2c), 25% E (23% E1b1b1a1, 2% E1b1b1b2a), 23% R (13% R1a, 10% R1b), 17% J (8% J2a, 7% J2b, 2% J1), 6% G2a, 1% H1a, 1% T[21]

A Bray–Curtis dissimilarity
B Chord distance
mtDNA
Complimentary evidence exists from mtDNA data. Bulgaria shows a very similar profile to other European countries – dominated by mitochondrial haplogroups Hg H (~42%), Hg U (~18%), Hg J/Hg T (~18%), and Hg K (~6%).[59] Like most Europeans, H1 is the prevailing subclade among Bulgarians.[60] Most of the U-carriers belong to U5 and U4. The distribution of the subclades of Haplogroup H have not been revealed. Recent studies show greater diversity within mt Haplogroups than once thought, as sub-haplogroups are being discovered, and often separate migrations and distributions of the Y-DNA haplogroups. While the Y-DNA variation in Europe is clinal, the mitochondrial is not.[61]
The results of the mitochondrial analyses find the Bulgarians more related to North Slavs. The general mtDNA analyses of the study find the Bulgarians in a cluster with Central Europeans, but others find them more related to Balkan peoples, or to both at the same time. According to the largest-scale mtDNA Bulgarian study involving 996 Bulgarian samples, comparing distribution of hgs H, H5, HV, HV0, R0a, J, U1, U2, U2e, U3, U4, U5a, U5b, U6, U7, U8, K, N1, N2, X, M, Т1, Т2, the Bulgarians came out nearest to the Poles, followed by Ukrainians, Croats, Czechs, while neighbouring Turks, Romanians and Greeks remained more or very distant.[62] Several other pan-European analyses of the same Bulgarian samples indicate that the neighbouring populations except the Macedonians are distant from the Bulgarians. According to these the Bulgarians are at minimal distance by mtDNA either to Hungarians, Székelys, Slovaks or to Macedonians, followed either by Ukrainians, Croats or Czechs, while neighbouring populations such as Turks, Romanians, Serbs and Greeks remain more distanced.[63][64][65] Italians also remain related according to these studies. According to another analysis, Bulgarians despite being part of a Slavic mitochondrial cluster, are most related to Romanians, followed by Hungarians and Czechs, but distant to Balkan Albanians and Bosniaks, it was also found out that Bulgarians are very distant to all Asian and African populations, but are least distanced by mtDNA to the Arkhangelsk Oblast in Russia[66] where also Y-DNA hg I2 prevails.[67] A more recent pan-Slavic mtDNA plot analyzing genetic distance, situates Bulgarians as sharing their position with Czechs, Romanians, Macedonians and Hungarians, while other close groups to them are Slovaks, Estonians and Latvians.[7] Others determined Albanians and Portuguese as most related to Bulgarians,[68] others determined northern Europeans and Slavs,[69] others Israeli[70] however some of these analyses depend on minimal variance that changes the proximal distance. Other studies consider the Bulgarians closest to Romanians, followed by Italians and Iberians, while Poles, Slovaks, and Czechs are determined more distanced.[71] According to other studies Romanians are the closest, followed by Hungarians, Greeks, Macedonians and then by Central Europeans.[72] It should be noted none of the analyses on genetic distance compares the subclades of Haplogroup H except for H5 due to lacking data, however the structure of the haplogroup of some central European, Balkan and other peoples may be similar to the Bulgarian, at least by division of older subclades of haplogroup H.
MtDNA haplgroups of ~1000 Bulgarians:[59]
- HV - 49%
- H - 41%
- H5 - 3%
- HV - 4%
- HV0 - 4%
- H - 41%
- U - 18%
- U1 - 1%
- U2e - 1%
- U3 - 2%
- U4 - 4%
- U5 - 8%
- U5a - 5%
- U5b - 3%
- U6 - 0%
- U7 - 1%
- U8 - <1%
- JT - 18%
- K - 6%
- N - 5%
- N1 - 3%
- N2 - 2%
- X - 2%
- M - 1%
- L - <1%
- R0a - <1%
- Others - <1%
auDNA and overall
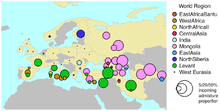
Whilst haploid markers such as mtDNA and Y-DNA can provide clues about past population history, they only represent a single genetic locus, compared to hundreds of thousands present in nuclear, autosomes. Although autosomal analyses often sample a small number of Bulgarians, by multiple autosomes multiple ancestral lines may be traced by an individual's 21 autosomes as opposed to one identical mtDNA or Y DNA sex chromosome, whose inheritance although clinal, demonstrates genetic drift often in statistics. Analyses of autosomal DNA markers gives the best approximation of overall 'relatedness' between populations, presenting a less skewed genetic picture compared to Y DNA haplogroups. This atDNA data shows that there are no sharp discontinuities or clusters within the European population. Rather there exists a genetic gradient, running mostly in a southeast to northwest direction. A study compared all Slavic nations and combined all lines of evidence, autosomal, maternal and paternal, including more than 6000 people for and at least 700 Bulgarians from previous studies, of which 13 were used for autosomal analysis (right image). The overall data situates the southeastern group (Bulgarians and Macedonians) in a cluster with Romanians, and they are at similar proximity to Gagauzes, Montenegrins and Serbs who are not part of another cluster but are described as 'in between' clusters.[7] Macedonians and Romanians consistingly keep being among most related to Bulgarians by au, mt, and Y-DNA[7] a conclusion backed also by a pan-European autosomal study investigating 500,568 SNP (loci) of 1,387 Europeans and including 1 or 2 Bulgarians,[73] other more or less extensive data sets situate Bulgarians and Romanians as their nearest .[74][75][76] Per HLA-DRB1 allele frequencies Bulgarians are also in a cluster with the same populations.[77] The Balto-Slavic study itself calculated genetic distance by SNP data of the multiple autosomes and ranked most proximal to Bulgarians the Serbs, followed by Macedonians, Montenegrins, Romanians, Gagauzes, Macedonian Greeks apart from Thessaloniki, the rest of the South Slavs, Hungarians, Slovaks, Czechs, and then by Greeks from Thessaloniki, Central Greece and Peloponese.[7] The East Slavs and Poles cluster together remaining less proximal to Bulgarians than Germans, among whom Slav admixture is also observed. Balts, however, according to the PCA analysis are less proximal to Bulgarians than Italians for example are.[7] Bulgarians are also only modestly close to their eastern neighbours – the Anatolian Turks, suggesting the presence of certain geographic and cultural barriers between them.[78] Despite various invasions of Altaic-speaking peoples in Europe, no significant impact from such Asian descent is recorded throughout southern and central Europe.[79]

The study claims that the major part of the Balto-Slavic genetic variation can be primarily attributed to the assimilation of the pre-existing regional genetic components, which differed for present West, East and South Slavic-speaking people.[7] For Slavic peoples correlations with linguistics came out much lower than high correlations with geography.[7] The South Slavic group, despite sharing a common language, is separated and has largely different genetic past from their northern linguistic relatives genetically.[4][7][12] Therefore, for the Bulgarians and most other South Slavs the most plausible explanation would be that their most sizable genetic components were inherited from indigenous Balkan pre-Slavic and pre-Bulgar population.[4][7][12] Another pan-Slavic Y-DNA study concludes that most of the Southern Slavic group is distinct from their Northern Slavic relatives, whose homogeneity on the other hand stretches form the Alps to Volga end even as far as the Pacific Ocean in Russia.[12] This means that there is a paternal genetic rather than a geographical factor separating these Slavic peoples. The South Slavs are characterized by featuring NRY hgs I2a and E plus 10% higher Mediterranean k2 autosomal component, while the Eastern and Western Slavs are characterized by the k3 component and hg R1a.[7] The current differentiation of high I-P37 and lower R-Z282 among South Slavs and vice versa among North Slavs suggests it was present prior to the Slavic settling in the Balkans as no relevant migrations occur later to change the frequencies.[7][80][81] The contribution of the Y chromosomes of peoples who had settled in the Balkans before the Slavic expansion is the most likely explanation of the phenomenon according to the other study on Y-haplotypes, concluded by its two separate analyses because of the complicity of the methods tracing the alleles.[12] The presence of two distinct genetic substrata in the genes of East-West and South Slavs would conclude that assimilation of indigenous populations by bearers of Slavic languages was a major mechanism of the spread of Slavic languages to the Balkan Peninsula.[7]
Southeastern Europeans share large numbers of common ancestors that date roughly to the times of the Slavic expansion around 1,500 years ago. The eastern European populations with high rates of (IBD) are highly coincident with the modern distribution of Slavic languages including Hungary, Romania, Greece and Albania, so it is speculated for Slavic expansion, anyway it was concluded that additional work and methods would be needed to verify this hypothesis. This study detects a considerable connection between Bulgarians and North Slavs that is the result of migrations no earlier than 1500 years ago.[76] A study on genetic admixture filtered to 474,491 autosomal SNPs and including 18 Bulgarians concluded that there is a recent excess of identical by descent sharing in Eastern Europe, and recent period of exchanged segments speculating that this may correspond to the Slavic expansion across this region. A signal at a low frequency among Balkan Slavs was detected that may have been inherited from the medieval Slavic settlers, but it was confirmed that this issue requires further investigation.[7] The short genetic distance of South Slavs does not extend to populations throughout the whole Balkan Peninsula and they are differentiated from all Greek sub-populations that are not Macedonian Greek.[7] The South Slavs share significantly fewer identical by descent segments for length classes with Greeks than with the group of East-West Slavs.[7] Most of the East-West Slavs share as many such segments with the South Slavs as they share with the inter-Slavic populations between them. This might suggest Slavic gene flow across the wide area and physical boundaries such as the Carpathian Mountains, including Hungarians, Romanians and Gagauz.[7] Notably, the number of common ancestors within the last 1,000 to 2,000 years is particularly high within eastern and Slavic-speaking Europe.[82] A high number of shared IBD segments among East Europeans that can be dated to around 1,000–2,000 YBP was revealed. The highest percentage of the total number of shared pairwise IBD segments is detected between the group of East-West Slavs and South Slavs (41% from the total number of IBD segments detected); Baltic speakers, Estonians (40%) and "inter-Slavic" Hungarians, Romanians and Gagauz (37%). East-West Slavs share these segments with Western Europeans (32%), Volga region populations (30) and North Caucasus (21%). South Slavs also share 41% with East-West Slavs and 37% with Inter-Slavic populations, they also share 31% with Western Europeans and 30% with Greeks. However, per one pair of individuals East-West Slavs share more IBD with Balts than with South Slavs, but not with the rest and the same amount with inter-Slavic as with South Slavs. Per one pair of individuals South Slavs keep sharing most IBD with East-South Slavs and the same amount with the inter-Slavic, followed by Greeks and Western Europeans.[7]
For the Bulgarians prevailing donor group in admixture with up to more than 40% are a northeastern group, consistent with the medieval Slav expansion, the date of the admixture event is set at 500-950 CE. The Slavic frequency of the Bulgarians is determined lower than that of Poles and Hungarians, and higher than that of Romanians and Greeks, roughly in between.[5] From data of participating groups, it is inferred that the event of admixture among Bulgarians in the Middle Ages is 46% Belarusian-like, of which most notably Lithuanian-like (23%) and Polish-like (19%), while another 50%+ being Cypriot-like, of which 14% Greek-like.[6] Some of the phenomena that distinguish western and eastern subgroups of the South Slavic people and languages can be explained by two separate migratory streams of different tribal groups of the future South Slavs via both: the west and east of the Carpathian Mountains.[8] By some assumed ancestral populations, about 50% of Bulgarians are in a category "North and East Europe" (right image) similar to the estimate above. It is higher in the West Balkans even among Montenegrins who have only 7% of Haplogroup R1a, which most likely means that the study combines together respective autosomes corresponding to I2a1b and R1a in this category. On the contrary of these estimates is the data for ancestry of 23andme, which separates Northeast European from Southeast European(Balkan) ancestry. In some 23andme examples most South Slavs and Romanians are on average put at over 60% of Balkan source, 10-20% of Northeastern European source, and a further 10% undetermined category "broadly European" that may reduce each frequency. In individuals from all over Romania, who typically plot with Bulgarians and share roughly the same amount of donor groups, Northeast European ancestry may range between several percents and over 45 per sampled individual, while the Balkan is an element with much higher lower and higher bound. The Romanian average per 46 samples of Northeast European ancestry came out 11%, but there is an additional amount of 10% "broadly European", <1% unassigned, which means that Northeast European category may be deflated by a maximum of 10%. Although these estimates are focused on regional sources rather than population sources, none of the two estimates does specifically estimate Slavic or Balto-Slavic admixture or specifies any dates of the regional sources. The Northeastern European percentage from 23andme samples is often by times less than the specified category by DNA.Land "North Slavic". Of the group several sampled Bulgarians by new methodology of DNA.Land the "North Slavic" element averages 25–30%.[83] For comparison, according to the latter data od the "Balkan" element is nonexistent in Belarusians, and most often does not occur in Poles and Ukrainians, but accounts for more than a third of the total in Croats and Bulgarians. Although their data is highly indicative, both 23andme and DNA.Land have some obvious scarce datasets showing most Cypriots as being of Italian ancestry.
The genetic diversity among Bulgarians is the reason of more inherited diseases[84] The blood type of 21,568 Bulgarians is 37% A+ , 28% 0+, 14% B+, 7% AB+, 6+ A-, 4% 0-, 2% B-, 1% AB-,[85] a distribution similar to the Sweden, the Czech Republic and Turkey.
Ancient DNA

Despite the most common haplogroup among Bulgarians is I2a1b at 20%, 8000 years old hunter-gatherer samples of the same haplogroup came out genetically very distant from Bulgarian and Balkan individuals by an autosomal analysis of skeletal remains from Loschbour cave in Luxembourg.[86]
Three out of four samples from Bulgarian Neolithic(6,500-5000 ybp) from Smyadovo came out mtDNA haplogroup H and the other one is T2e, while another 5,500-4,000 old sample from Durankulak is U52a2. Several mtDNA Bulgarian samples considered part of the Yamna culture came out haplogroups H, T2a1b1a, U2e1a, U5a1 and K.[39]
Computing the frequency of common point mutations of several mtDNA Thracian remains from Romania with haplogroups H17, H22 and HV has resulted that the Italian (7.9%), the Albanian (6.3%) and the Greek (5.8%) have shown a bias of closer genetic kinship with the Thracian individuals than the Romanian and Bulgarian individuals (4.2%) , but it was noted that more mtDNA sequences from Thracian individuals are needed in order to perform a complex objective statistical analysis.[87] From seven Thracian samples aged about 3 millenia from Gabova Mogila and Shekerdja Mogila in Sliven Province, and from Bereketska Mogila in Stara Zagora Province, two were identified as belonging to mtDNA Haplogroup D, presumably associated with East Asia. Haplogroup W5a was found among two individuals and H1an2. H14b1 was also found.[88] Four samples from Iron Age Bulgaria were studied, the official study confirmed only that the two are male and mtDNA of two individuals - U3b for the Svilengrad man and HV for the Stambolovo individual. Haplogroups U for the Krushare man, U2e for the Vratitsa individual have been identified. Those individuals were from Thracian burial sites and are dated at around 450-1500 BC.[89] Unofficial analysis of the raw data claims that the first one is positive for Y-DNA Haplogroup E-Z1919 or H-Z14031 (H1b1). It also claims that according to the SNPs all the four samples came out male and also in the man from Krushare Haplogroup J-PF5197 (J2a1a1a1b2) was found, while another man's haplogroup came out negative for E, I and J and remained unknown but is likely R1.[90][91] According to an autosomal analysis of DNA Land, the Svilengrad man came out 100% Mediterranean islander,[92] while the Stambolovo man appears to be 99% Balkan.[93] One of the suprising results are of 3,500-3,100 years old samples from Vratitsa, which came out 60% Northwest European 24% Southwestern European(22% Sardinian),5% Ashkenazi, 5% Mbuti, 3% Native American, 3% South/Central European, 1% North Slavic,[94] along with a 2,400 years old sample from Krushare that is 32% Southwestern European, 26% Northwestern European, 26% Balkan, 5% Central Indo European, 3% Mbuti, 3% Finnish, 3% North Slavic, 1% Ambiguous, 1% Amazonian,[95] in comparison a sample from Iron Age Montenegro is suprising in the opposite direction and came out 64% North Slav and 50% Yamnaya.[96] For the man from Krushare the authors explicitly stated "However, the DNA damage pattern of this individual does not appear to be typical of ancient samples, indicating a potentially higher level of modern DNA contamination.". While the Svilengrad man still shows the highest proportion of Sardinian ancestry, the Krushare more resembles the hunther-gatherer individuals.
20 samples from medieval Bulgarian sites were alleged as originally Bulgar, but there is no evidence for that.[97] They were from a burial site from the Monastery of Mostich in Preslav, Nozharevo, Tuhovishte and most came out European mtDNA haplogroup H, including H1, H1an2, H1r1, H1t1a1, H2a2a1 H5, H13a2c1, H14b1, HV1, J, J1b1a1, T, T2, U4a2b, U4c1 and U3 with the half belonging to Haplogroup H. It was shown a short genetic distance between these samples and modern Bulgarians.[98]
After at least 20 mediaval(10-14th century) mtDNA samples from Cedynia and Lednica in Poland, possibly Slavic, had been studied, the 855 sampled modern Bulgarians come out overally the closest group to these samples out of 20 other European nations and moreover, they share the highest value of haplotypes with the medieval Polish population more than any other compared nation does. Those medieval haplogroups included H, H1a, K1, K2, X2, X4, HV, J1b, R0a, HV0, H5a1a, N1b, T1a, J1b and W. The samples came out distant from modern Polish population, but nearest to the modern Bulgarian and Czech population.[64] 20 medieval(9-12th century) samples from Slovakian sites Nitra Šindolka and 8 from Čakajovce were compared to modern population and Bulgarians, and Portuguese came out nearest to them by genetic distance, however all these came out distant to modern Slovak population.[71]

Further evidence from ancient DNA, reconsiderations of mutation rates, and collateral evidence from autosomal DNA growth rates suggest that the major period of European population expansion occurred after the Holocene. Thus the current geographic spread and frequency of haplogroups has been continually shaped from the time of Palaeolithic colonization to beyond the Neolithic.[99] This process of genetic shaping continued into recorded history, such as the Slavic migrations.[100]
Recent studies of ancient DNA have revealed that European populations are largely descending from three ancestral groups. The first one are Paleolithic Siberians, the second one are Paleolithic European hunter-gatherers, and the third one are early farmers and later arrivals from the Near East and West Asia. According to this, Bulgarians are predominantly (~ 2/3) descending from early Neolithic farmers spreading the agriculture from Anatolia, and from West Asian Bronze Age invaders and cluster together with other Southern Europeans. Another of the admixture signals in that farmers involves some ancestry related to East Asians, with ~ 2% total Bulgarian ancestry proportion linking to a presence of nomadic groups in Europe, from the time of the Huns to that of the Ottomans. A third signal involves admixture between the North European group from one side and the West Asian - Early farmers' group from another side, at approximately the same time as the East Asian admixture, ca. 850 AD. This event may correspond to the expansion of Slavic language speaking people. The analysis documents the hunter-gatherer admixture in Bulgarians at a level from ca. 1/3.[101] The impact of Yamnaya culture is estimated at 20-30%, which is most common among the Slavs.
According to Genographic Project's autosomal study called Your Regional Ancestry based on nine regional affiliations, the Bulgarians regional ancestry results are as follows: 47% Mediterranean and 20% southwest Asian impact, which reflect the strong influence of neolithic agriculturalists from the Fertile Crescent; 31% Northern European component that reflects Paleolithic hunter-gathers' ancestry; 2% Northeast Asian component which shows there have been some mixings with Asiatic invaders.[102]
Physical features
According to early 20th century statistics on 230,000 Bulgarians, between 9-12% of them were blond-type depending on the region, the brunette type makes up 42-47%, while 43-46% are determined as an intermediate type (light eyes and dark hair or vice versa).[103] The highest proportion of 12% blond type is placed in southwestern Bulgaria, followed by the southern provinces Macedonia, Thrace and least of 9% is in northern Bulgaria. The same and other nationwide studies observe heterogeneity in cephalic index in northern (brachycephalic) and southern (mesocephalic) Bulgaria.[104] Results from a 20th century study on 9,000 persons of Bulgarian ethnicity at an average age of 22, including about 400 Muslim Bulgarians and 500 from Vardar Macedonia concluded that 13% of the males are blond-haired and light-eyed at the same time by Martin-Saller scale, while 55% of them are brown/black-haired and dark-eyed.[105] Other statistics consider 13% as blue-eyed and 25% as grey-eyed.[106]
A late 20th century anthropological study[107] on over 5,000 adults, involving only participants aged 30–40 and Romani participants, other minorities and dark-skinned people in the common statistics of unknown number, found relatively homogeneous results by regions. It involved also numerous researches. One of them found the skin color "white" to be prevailing among all genders and regions with 80%, but some 20% were determined as mat and light mat skinned. Consistency of more frequent natural light hair was found among women, and slightly more frequent light eyes among men. A hair color "brown-black" (80% of men, 67% women) was found prevailing among all genders and regions. By Fischer-Saller scale 3% of the men were determined blond, mostly dark blonde, and 18% brown-haired. 7% of the sampled women were determined as naturally blonde and 26% as brown-haired. Most frequent naturally blonde women were found in Sofia-city, namely 12% of the women there, of which 5% were light-blonde. Reddish-haired men and women make up 0.1 and 0.2%, peaking at almost a percent in the former Haskovo and Burgas provinces. Eye color by Martin distribution found most often prevailing "dark, black-brown and brown" color (48% men, 57% women). In some regions "motley color - green-brown, gray-brown, blue-brown" color (45% men, 38% women) was found leading, peaking among Lovech men at 50%. The gray and blue eyes were detected at 6% among the women and 8% among the men. Most of the women have moderately prominent cheekbones (60%) and sometimes strongly (14%), men have the same characteristics at frequencies 40% and 3%. 77 and 65 kg is the mean weight of men and women and the BMI - 26. 45 is the mean hand strength. The hell/dark dental colour of women and men is 21%/16% and 49%/2% respectively, the rest being a middle color. Straight nose occurs at 70% of both genders, convex nose at 28%/21% of men/women and concave nose at 3%/8%, while only in Haskovo the concave nose among women (11%) outnumbers the convex (10%). Without a change of more than a centimeter for the past century, 171/159 cm was the average male/female height and 173/161 cm in Sofia. 52% of the men recorded were between 170–178 cm and 8% taller than this, while the highest of all was 191 cm. 41% of the recorded women were 159–168 cm and 6% - taller than this.
More recently it was estimated that 175/163 cm is the average male/female height and 178/164 cm in Sofia.[108]
See also
External links
References
- ↑ One Europe, many nations: a historical dictionary of European national groups, James Minahan, Greenwood Publishing Group, 2000, ISBN 0-313-30984-1, pp. 134–135. Google Books. 2000-01-01. ISBN 9780313309847. Retrieved 2011-11-13.
- ↑ Fine, John Van Antwerp (1991). The early medieval Balkans: a critical survey from the sixth to the late twelfth century. University of Michigan Press. p. 308. ISBN 978-0-472-08149-3.
- ↑ Kopeček, Michal (2007). Balázs Trencsényi, ed. Discourses of collective identity in Central and Southeast Europe (1770–1945): texts and commentaries. Central European University Press. p. 240. ISBN 978-963-7326-60-8.
- 1 2 3 Expansions: Competition and Conquest in Europe Since the Bronze Age, Reykjavíkur Akademían, 2010, ISBN 9979992212, p. 194.
- 1 2 3 Garrett Hellenthal et al A genetic atlas of human admixture history
- 1 2
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 Genetic Heritage of the Balto-Slavic Speaking Populations: A Synthesis of Autosomal, Mitochondrial and Y-Chromosomal Data, Alena Kushniarevich et al. September 2, 2015; doi:10.1371/journal.pone.0135820
- 1 2 The Slavic Languages, Roland Sussex, Paul Cubberley, Publisher Cambridge University Press, 2006, ISBN 1139457284, p. 42.
- ↑ Hupchick, Dennis P. The Balkans: From Constantinople to Communism. Palgrave Macmillan, 2004. ISBN 1-4039-6417-3
- 1 2 3
- ↑ Karas, Haliny. Gwary polskie. Przewodnik multimedialny. ISBN 978-83-62844-09-8.
- 1 2 3 4 5 6 7 Rebala et. al. (2007) Y-STR variation among Slavs: evidence for the Slavic homeland in the middle Dnieper basin
- ↑ Science, 14 February 2014, Vol. 343 no. 6172, p. 751, A Genetic Atlas of Human Admixture History, Garrett Hellenthal at al.: " CIs. for the admixture time(s) overlap but predate the Mongol empire, with estimates from 440 to 1080 CE (Fig.3.) In each population, one source group has at least some ancestry related to Northeast Asians, with ~2 to 4% of these groups total ancestry linking directly to East Asia. This signal might correspond to a small genetic legacy from invasions of peoples from the Asian steppes (e.g., the Huns, Magyars, and Bulgars) during the first millennium CE."
- ↑ Y-Chromosome Diversity in Modern Bulgarians: New Clues about Their Ancestry, Karachanak S, Grugni V, Fornarino S, Nesheva D, Al-Zahery N, et al. (2013) Retrieved Oct 2013.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Karachanak 2013
- ↑ Varzari 2003 and 2007, Kushniarevich 2015, Noevski 2010, Marjanovic 2005, Mrsic 2012, Todorovic 2014, Vakar et al 2010
- 1 2 Jean Manco. "Mesolithic Western Eurasian DNA".
- ↑ "Peopling of Europe 2014 – Identifying the Ghost Population". DNAeXplained - Genetic Genealogy.
- ↑ Rootsi 2004
- 1 2 yfull
- 1 2 3 4 5 6 7 8 9 "Family Tree DNA - Bulgarian DNA Project".
- ↑ "L621".
- ↑ "Славяне и субстрат".
- 1 2 (Karachanak 2013)
- 1 2 Cruciani 2004
- ↑ "Tracing Past Human Male Movements in Northern/Eastern Africa and Western Eurasia: New Clues from Y-Chromosomal Haplogroups E-M78 and J-M12" (PDF).
- ↑ Lacan 2011
- ↑ Cruciani
- ↑ Sarno et al 2015, King et al 2008, Mirabal et al, Petricic et al 2005, Martinez-Cruz 2012
- ↑ Kayser 2005
- 1 2 "European Journal of Human Genetics - Supplementary Information for article: The phylogenetic and geographic structure of Y-chromosome haplogroup R1a".
- ↑ Martinez-Cruz 2001
- ↑
- ↑
- ↑
- ↑
- ↑
- ↑ Cruciani 2010, Klyosov 2012
- 1 2 Manco, Jean (29 December 2015). "Ancient Western Eurasian DNA of the Copper and Bronze Ages". ancestraljourneys.org. Retrieved 1 May 2016.
- ↑ Myles et al
- ↑ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3259916/
- ↑ Marjanovic 2005 et al, Volgi 2008 et al, Martinez Cruz 2012 et al
- ↑ Cinniouglu et al
- ↑ Rosser, ZH; Zerjal, T; Hurles, ME; et al. (December 2000). "Figure 5: Y-Chromosomal Diversity in Europe Is Clinal and Influenced Primarily by Geography, Rather than by Language". The American Journal of Human Genetics. 67: 1526–43. doi:10.1086/316890. PMC 1287948
 . PMID 11078479.
. PMID 11078479. - ↑ Martinez-Cruz, B; Ioana, M; Calafell, F; Arauna, LR; Sanz, P; Ionescu, R; Boengiu, S; Kalaydjieva, L; Pamjav, H; Makukh, H; Plantinga, T; van der Meer, JW; Comas, D; Netea, MG (2012). "Y-Chromosome Analysis in Individuals Bearing the Basarab Name of the First Dynasty of Wallachian Kings". PLoS ONE. 7: e41803. doi:10.1371/journal.pone.0041803. PMC 3404992
 . PMID 22848614.
. PMID 22848614. - ↑ Kasperaviciūte, D; Kucinskas, V; Stoneking, M (September 2004). "Y Chromosome and Mitochondrial DNA Variation in Lithuanians". Annals of Human Genetics. 68: 438–452. doi:10.1046/j.1529-8817.2003.00119.x. PMID 15469421.
- ↑ Karachanak 2013, Table 2
- ↑ Völgyi et al 2008
- ↑ Vágó-Zalán Andrea 2012 A magyar populáció genetikai elemzése nemi kromoszómális markerek alapján.
- ↑ Arnaiz-Villena; et al. (1992). "(June 2003) HLA Genes in the Chuvashian Population from European Russia: Admixture of Central European and Mediterranean Populations". Human Biology. 75 (3): 375–392. doi:10.1353/hub.2003.0040. PMID 14527201.
- ↑ Jakovski; et al. (2011). "Genetic data for 17 Y-chromosomal STR loci in Macedonians in the Republic of Macedonia". Forensic Sci. Int. Genet. 5: e108–e111. doi:10.1016/j.fsigen.2011.04.005.
- ↑ Alshamali et al. Local Population Structure in Arabian Peninsula Revealed by Y-STR Diversity
- ↑ Mrsic et al
- ↑ Varzari 2006
- ↑
- ↑ Karachanak 2009
- ↑ "Bulgarian_Y_Table". Google Docs.
- ↑ Martinez-Cruz 2012
- 1 2 Karachanak 2012
- ↑ "Family Tree DNA - Bulgarian DNA Project".
- ↑ Rosser et. al. Y-Chromosomal Diversity in Europe Is Clinal and Influenced Primarily by Geography, Rather than by Language
- ↑ Sena Karachanak et al. "Bulgarians vs the other European populations: a mitochondrial DNA perspective." International Journal of Legal Medicine, June 15, 2011.
- ↑ Mitochondrial DNA Suggests a Western Eurasian origin for Ancient (Proto-) Bulgarians
- 1 2 p. 100 Anna Juras, Etnogeneza Słowian w świetle badań kopalnego DNA, Praca doktorska wykonana w Zakładzie Biologii Ewolucyjnej Człowieka Instytutu Antropologii UAM w Poznaniu pod kierunkiem Prof. dr hab. Janusza Piontka
- ↑ mtDNA PCA
- ↑ balanovsky 2012
- ↑ "PubMed Central Image Viewer.".
- ↑
- ↑
- ↑ Wolfgang Haak et al.
- 1 2 Csákyová, V; Szécsényi-Nagy, A; Csősz, A; Nagy, M; Fusek, G; Langó, P; Bauer, M; Mende, BG; Makovický, P; Bauerová, M (2016). "Maternal Genetic Composition of a Medieval Population from a Hungarian-Slavic Contact Zone in Central Europe". PubMed Central (PMC). 11: e0151206. doi:10.1371/journal.pone.0151206. PMC 4786151
 . PMID 26963389.
. PMID 26963389. - ↑ The mitochondrial DNA makeup of Romanians: A forensic mtDNA control region database and phylogenetic characterization
- ↑ John Novembre et al. Genes mirror geography within Europe, Nature, 6 November 2008, 456 (7218):98-101.
- ↑
- ↑ "Ancestry Composition AU, DE, FR & EU - 23andMe".
- 1 2 PLOS —The Geography of Recent Genetic Ancestry across Europe, Peter Ralph, Graham Coo, May 7, 2013 – doi:10.1371/journal.pbio.1001555
- ↑ "Greeks - Sub-Saharan Africa - Origin - Arnaiz-Villena".
- ↑ Novembre 2008, Yanusbaev 2012.
- ↑ Iosif Lazaridis et al. Ancient human genomes suggest three ancestral populations for present-day Europeans, bioRxiv doi:10.1101/001552
- ↑ Karmin M 2015
- ↑ Underhill 2014
- ↑
- ↑ http://dnagenealogy-bg.org/mybb/showthread.php?tid=425&page=3
- ↑ Two autosomal recessive neuromuscular disorders – FRDA and LGMD2A, in a single Bulgarian pedigree, Savina Tincheva et al. Scripta Scientifica Medica Vol 47, No 4 (2015). DOI
- ↑
- ↑ Ancient human genomes suggest three ancestral populations for present-day Europeans, Iosif Lazaridis et al., Nature 513, 409–413 (18 September 2014) doi:10.1038/nature13673
- ↑ G. Cardos, V. Stoian, N. Miritoiu, A. Comsa, A. Kroll, S. Voss, and Alexander Rodewald. "Paleo-mtDNA analysis and population genetic aspects of old Thracian populations from South-East of Romania." Romanian Society of Legal Medicine 12:4 (2004): pages 239-246.
- ↑
- ↑ Carpenter, ML; Buenrostro, JD; Valdiosera, C; Schroeder, H; Allentoft, ME; Sikora, M; Rasmussen, M; Gravel, S; Guillén, S; Nekhrizov, G; Leshtakov, K; Dimitrova, D; Theodossiev, N; Pettener, D; Luiselli, D; Sandoval, K; Moreno-Estrada, A; Li, Y; Wang, J; Gilbert, MT; Willerslev, E; Greenleaf, WJ; Bustamante, CD (2013). "Pulling out the 1%: Whole-Genome Capture for the Targeted Enrichment of Ancient DNA Sequencing Libraries". Am. J. Hum. Genet. 93: 852–64. doi:10.1016/j.ajhg.2013.10.002. PMC 3824117
 . PMID 24568772.
. PMID 24568772. - ↑ Genetiker (1 September 2015). "More Y-SNP calls from Iron and Bronze Age Bulgaria". genetiker.wordpress.com. Retrieved 1 May 2016.
- ↑ Genetiker (11 June 2014). "Analyses of Iron and Bronze Age Bulgarian genomes". genetiker.wordpress.com. Retrieved 1 May 2016.
- ↑ http://i.imgur.com/ccwoGdC.png
- ↑ http://i.imgur.com/OJOONYw.png
- ↑ http://i.imgur.com/SLBoAu4.png
- ↑ http://i.imgur.com/biGCIWY.png
- ↑ http://i.imgur.com/hIRTJHx.png
- ↑ Божидар Димитров за изследване на БАН: Копелета, има пари за усвояване! 11.10.2013 г. Новини.Бг.
- ↑ Mitochondrial DNA Suggests a Western Eurasian origin for Ancient (Proto-) Bulgarians, (2015). Human Biology Open Access Pre-Prints. Paper 69. http://digitalcommons.wayne.edu/humbiol_preprints/69
- ↑ Pinhasi 2012, Ricaut 2012.
- ↑ Rower 2005, Ralph 2012
- ↑ Science 14 February 2014, Vol. 343 no. 6172 pp. 747-751, "A Genetic Atlas of Human Admixture History", Garrett Hellenthal at al.
- ↑ National Genographic project Your Regional Ancestry: Reference Populations.
- ↑ Popov, Metodiy
- ↑
- ↑ акад. Методий Попов - Антропология на българския народ, том - I , Физически облик на българите, БАН, София, 1959, pp. 261
- ↑ Race & History. Routledge. ISBN 9781317847137.
- ↑ Yordanov, Yordan. Anthropology of the Bulgarian Population at the End of the 20th Century (30-40 Years old persons). Bulgarian Academy of Sciences, Sofia, 2006. Professor Marin Drinov Publishing House. p. 218, 219, 220, 223, 224, 406, 412
- ↑