General bacterial porin family
| Gram-negative porin | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| Symbol | Porin_1 | ||||||||
| Pfam | PF00267 | ||||||||
| Pfam clan | CL0193 | ||||||||
| InterPro | IPR001702 | ||||||||
| PROSITE | PDOC00498 | ||||||||
| SCOP | 1mpf | ||||||||
| SUPERFAMILY | 1mpf | ||||||||
| TCDB | 1.B.1 | ||||||||
| OPM superfamily | 31 | ||||||||
| OPM protein | 1pho | ||||||||
| CDD | cd01345 | ||||||||
| |||||||||
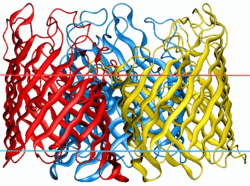
General bacterial porins are a family of proteins from the outer membranes of Gram-negative bacteria. The porins act as molecular filters for hydrophilic compounds.[1] They are responsible for the 'molecular sieve' properties of the outer membrane. Porins form large water-filled channels which allow the diffusion of hydrophilic molecules into the periplasmic space. Some porins form general diffusion channels that allow any solute up to a certain size (that size is known as the exclusion limit) to cross the membrane, while other porins are specific for one particular solute and contain a binding site for that solute inside the pores (these are known as selective porins). As porins are the major outer membrane proteins, they also serve as receptor sites for the binding of phages and bacteriocins.
General diffusion porins usually assemble as a trimer in the membrane, and the transmembrane core of these proteins is composed exclusively of beta strands.[2] It has been shown[3] that a number of porins are evolutionarily related, and these porins are:
- E.Coli PhoE phosphoporin (Phosphate anion selective TC# 1.B.1.1.2), OmpC(TC# 1.B.1.1.3) OmpF(general porins of differing pore diameter TC# 1.B.1.1.1), NmpC (TC# 1.B.1.1.4 anion selective, transporting a variety of drugs and small neutral compounds)
- Bacteriophage PA.2 LC Porin(TC# 1.B.1.1.5) (not well characterized).
- Neisseria PorA(TC# 1.B.1.5.2), PorB(TC# 1.B.1.5.4)
Structure of Porins
Porins are composed of β-strands, which are, in general, linked together by beta turns on the periplasmic side of the outer membrane and long loops on the external side of the membrane. The β strand lie in an antiparallel fashion and form a cylindrical tube, called a β-barrel[2]. The amino acid composition of the porin β-strands are unique in that polar and non-polar residues alternate along them. This means that the non-polar residues face outwards so as to interact with the non-polar lipid membrane, whereas the polar residues face inwards into the center of the β-barrel to form the aqueous channel. The phospholipids that comprise the outer membrane give it the same semi-permeable characteristics as the cytoplasmic membrane[4]
The porin channel is partially blocked by a loop, called the eyelet, which projects into the cavity. In general, it is found between strands 5 and 6 of each barrel, and it defines the size of solute that can traverse the channel. It is lined almost exclusively with charged amino acyl residues arranged on opposite sides of the channel, creating a transversal electric field across the pore. The eyelet has a local surplus of negative charges from four glutamic acid and seven aspartic acid residues (in contrast to one histidine, two lysine and three arginine residues) is partially compensated for by two bound calcium atoms, and this asymmetric arrangement of molecules is thought to have an influence in the selection of molecules that can pass through the channel[3].
Homologous Families
Three dimensional structural analyses show that there are many(at-least 48) other families which share sufficient sequence similarity to the General Bacterial Porin(GBP) family. are homologous in structure and function to General bacterial porin family. One such family is The Sugar Porin (SP) Family. (TC# 1.B.3) The SP family includes the well characterized maltoporin of E. coli for which the three-dimensional structures with and without its substrate have been obtained by X-ray diffraction. The protein consists of an 18 β-stranded β-barrel in contrast to proteins of the general bacterial porin family (GBP) and the Rhodobacter PorCa Porin (RPP) family(TC# 1.B.7)) which consist of 16 β-stranded β-barrels. Although maltoporin contains a wider beta-barrel than the porins of the GBP (TC# 1.B.1) and RPP families(TC# 1.B.7), it exhibits a narrower channel, showing only 5% of the ionic conductance of the latter porins.
The Rhodobacter PorCa Protein, the only well characterized member of the RPP family, was the first porin to yield its three-dimensional structure by X-ray crystallography. It has a 16-stranded β-barrel structure similar to that of the members of the GBP (TC #1.B.1) family. Paupit et al. (1991) presented crystal structures of phosphoporin (PhoE; TC# 1.B.1.1.2), maltoporin (LamB; TC# 1.B.3.1.1) and Matrixporin (OmpF), all of E. coli, and found these have 3-d folds similar to that of the Rhodobacter porin, PorCa. Structural and sequence analysis provide firm evidence that the GBP, SP and RPP families together with 44 additional families in TCDB belong to a single superfamily. However, we have been able to demonstrate homology between members of families GBP and RPP using statistical means (M. Saier, unpublished results).
Porin Superfamilies
General bacterial porin family belongs to Porin Superfamily I. The homologous families Sugar Porin(SP) family and Rhodobacter PorCa Porin (RPP) Family also belong to the Porin Superfamily I.
Subfamilies
References
- ↑ Benz R, Bauer K (1988). "Permeation of hydrophilic molecules through the outer membrane of gram-negative bacteria. Review on bacterial porins". Eur. J. Biochem. 176 (1): 1–19. doi:10.1111/j.1432-1033.1988.tb14245.x. PMID 2901351.
- ↑ Jap BK, Walian PJ (1990). "Biophysics of the structure and function of porins". Q. Rev. Biophys. 23 (4): 367–403. doi:10.1017/S003358350000559X. PMID 2178269.
- ↑ Pattus F, Jeanteur D, Lakey JH (1991). "The bacterial porin superfamily: sequence alignment and structure prediction". Mol. Microbiol. 5 (9): 2153–2164. doi:10.1111/j.1365-2958.1991.tb02145.x. PMID 1662760.
- ↑ Van Gelder P, Saint N, van Boxtel R, Rosenbusch JP, Tommassen J (1997). "Pore functioning of outer membrane protein PhoE of Escherichia col". Protein Eng. 10 (6): 699–706. doi:10.1093/protein/10.6.6. PMID 9278284.