Gene redundancy
Gene redundancy is the existence of multiple genes in the genome of an organism that perform the same function. This is the case for many sets of paralogous genes. When an individual gene in such a set is disrupted by mutation or targeted knockout, there can be little effect on phenotype as a result of gene redundancy, whereas the effect is large for the knockout of a gene with only one copy.[1]
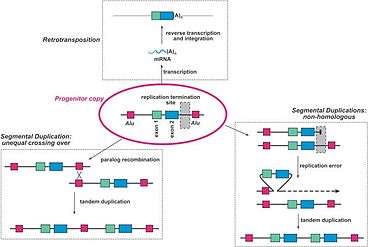
Origin and Evolution of Redundant Genes
Gene redundancy most often results from Gene duplication. When a gene is duplicated within a genome, the two copies are initially functionally redundant.[2] Three of the more common mechanisms of gene duplication are retroposition, unequal crossing over, and non-homologous segmental duplication. Retroposition is when the mRNA transcript of a gene is reverse transcribed back into DNA and inserted into the genome at a different location. During unequal crossing over, homologous chromosomes exchange uneven portions of their DNA. This can lead to the transfer of one chromosome's gene to the other chromosome, leaving two of the same gene on one chromosome, and no copies of the gene on the other chromosome. Non-homologous duplications result from replication errors that shift the gene of interest into a new position. A tandem duplication then occurs, creating a chromosome with two copies of the same gene. Figure 1 to the left provides a good visual explanation of these three mechanisms.[3]

As the genome is replicated over many generations, the redundant gene's function will most likely evolve due to Genetic drift. This means that the gene will acquire mutations that change the overall function. The three mechanisms of functional differentiation in genes are nonfunctionalization (or gene loss), neofunctionalization and subfunctionalization.[4]
During nonfunctionalization, or degeneration/gene loss, one copy of the duplicated gene acquires mutations that render it inactive or silent. At this time, the gene has no function and is called a pseudogene. Pseudogenes can be lost over time due to genetic mutations. Neofunctionalization occurs when one copy of the gene accumulates mutations that give the gene a new, beneficial function that is different than the original function. Subfunctionalization occurs when both copies of the redundant gene acquire mutations. Each copy becomes only partially active; two of these partial copies then act as one normal copy of the original gene. Figure 2 to the right provides a visual explanation.
Gene Maintenance Hypotheses
The evolution and origin of redundant genes remain unknown, largely because evolution happens over such a long period of time. Theoretically, a gene can not be maintained without mutation unless it has a selective pressure acting on it. Gene redundancy, therefore, would allow both copies of the gene to accumulate mutations as long as the other was still able to perform its function. This means that all redundant genes should theoretically become a pseudogene and eventually be lost. Scientists have devised two hypothesis as to why redundant genes can remain in the genome: the backup hypothesis and the piggyback hypothesis.[5]
The backup hypothesis proposes that redundant genes remain in the genome as a sort of "back-up plan". If the original gene loses its function, the redundant gene is there to take over and keep the cell alive. The piggyback hypothesis states that two paralogs in the genome have some kind of non-overlapping function as well as the redundant function. In this case, the redundant part of the gene remains in the genome due to the proximity to the area that codes for the unique function.[6] The reason redundant genes remain in the genome is an ongoing question and gene redundancy is being studied by researchers everywhere. There are many hypotheses in addition to the backup and piggyback models. For example, at the University of Michigan, a study provides the theory that redundant genes are maintained in the genome by reduced expression.
Research
Gene Families and Phylogeny
Researchers often use the history of redundant genes in the form of gene families to learn about the phylogeny of a species. It takes time for redundant genes to undergo functional diversification; the degree of diversification between orthologs tells us how closely related the two genomes are. Gene duplication events can also be detected by looking at increases in gene duplicates.
A good example of using gene redundancy in evolutionary studies is the Evolution of the KCS gene family in plants. This paper studies how one KCS gene evolved into an entire gene family via duplication events. The number of redundant genes in the species allows researchers to determine when duplication events took place and how closely related species are.
Locating and Characterizing Redundant Genes
Currently, there are three ways to detect paralogs in a known genomic sequence: simple homology (FASTA), gene family evolution (TreeFam) and orthology (eggNOG v3).[7] Since the Human Genome Project's completion, researchers are able to annotate the human genome much more easily. Using online databases like the Genome Browser at UCSC, researchers can look for homology in the sequence of their gene of interest.
Human Redundant Genes
Olfactory Receptors
The Human Olfactory Receptor (OR) gene family contains 339 intact genes and 297 pseudogenes. These genes are found in different locations throughout the genome, but only about 13% are on different chromosomes or on distantly spaced loci. 172 subfamilies of OR genes have been found in humans, each at its own loci. Because the genes in each of these subfamilies are structurally and functionally similar, and in close proximity to each other, it is hypothesized that each evolved from single genes undergoing duplication events. The high number of subfamilies in humans explains why we are able to recognize so many odors.
Human OR genes have homologues in other mammals, such as mice, that demonstrate the evolution of Olfactory Receptor genes. One particular family that is involved in the initial event of odor perception has been found to be highly conserved throughout all of vertebrate evolution.[8]
Disease
Duplication events and redundant genes have often been thought to have a role in some human diseases. Large scale whole genome duplication events that occurred early in vertebrate evolution may be the reason that human monogenic disease genes often contain a high number of redundant genes. Chen et al. hypothesizes that the functionally redundant paralogs in human monogenic disease genes mask the effects of dominant deletorious mutations, thereby maintaining the disease gene in the human genome.[9]
Whole genome duplications may be a leading cause of retention of some tumor causing genes in the human genome.[10] For example, Strout et al.[11] have shown that tandem duplication events, likely via homologous recombination, are linked to acute myeloid leukemia. The partial duplication of the ALL1 (MLL) gene is a genetic defect has been found in patients with acute myeloid leukemia.
References
- ↑ Pérez-Pérez JM, Candela H, Micol JL (August 2009). "Understanding synergy in genetic interactions". Trends Genet. 25 (8): 368–76. doi:10.1016/j.tig.2009.06.004. PMID 19665253.
- ↑ Wagner, Andreas (1996). "Genetic Redundancy Caused By Gene Duplications And Its Evolution In Networks Of Transcriptional Regulators". Biological Cybernetics.
- ↑ Hurles, Matthew. "Gene Duplication: The Genomic Trade in Spare Parts". PLoS Biology. 2 (7). doi:10.1371/journal.pbio.0020206. PMC 449868
 . PMID 15252449.
. PMID 15252449. - ↑ Lynch, M. (2000). "The Evolutionary Fate and Consequences of Duplicate Genes". Science.
- ↑ Zhang, Jianzhi (2012). "Genetic Redundancies and Their Evolutionary Maintenance". Evolutionary Systems Biology Advances in Experimental Medicine and Biology.
- ↑ Qian, Wenfeng; Liao, Ben-Yang; Chang, Andrew Y.-F.; Zhang, Jianzhi (2010-10-01). "Maintenance of duplicate genes and their functional redundancy by reduced expression". Trends in genetics : TIG. 26 (10): 425–430. doi:10.1016/j.tig.2010.07.002. ISSN 0168-9525. PMC 2942974
 . PMID 20708291.
. PMID 20708291. - ↑ Bina, Minou (2006-01-01). "Identification and mapping of paralogous genes on a known genomic DNA sequence". Methods in Molecular Biology (Clifton, N.J.). 338: 21–29. doi:10.1385/1-59745-097-9:21. ISSN 1064-3745. PMID 16888348.
- ↑ Malnic, Bettina; Godfrey, Paul A.; Buck, Linda B. (2004-02-24). "The human olfactory receptor gene family". Proceedings of the National Academy of Sciences of the United States of America. 101 (8): 2584–2589. doi:10.1073/pnas.0307882100. ISSN 0027-8424. PMC 356993
 . PMID 14983052.
. PMID 14983052. - ↑ Chen, Wei-Hua; Zhao, Xing-Ming; van Noort, Vera; Bork, Peer (2013-05-01). "Human Monogenic Disease Genes Have Frequently Functionally Redundant Paralogs". PLoS Computational Biology. 9 (5). doi:10.1371/journal.pcbi.1003073. ISSN 1553-734X. PMC 3656685
 . PMID 23696728.
. PMID 23696728. - ↑ Malaguti, Giulia; Singh, Param Priya; Isambert, Hervé (2014-05-01). "On the retention of gene duplicates prone to dominant deleterious mutations". Theoretical Population Biology. 93: 38–51. doi:10.1016/j.tpb.2014.01.004. ISSN 1096-0325. PMID 24530892.
- ↑ Strout, Matthew P.; Marcucci, Guido; Bloomfield, Clara D.; Caligiuri, Michael A. (1998-03-03). "The partial tandem duplication of ALL1 (MLL) is consistently generated by Alu-mediated homologous recombination in acute myeloid leukemia". Proceedings of the National Academy of Sciences of the United States of America. 95 (5): 2390–2395. doi:10.1073/pnas.95.5.2390. ISSN 0027-8424. PMC 19353
 . PMID 9482895.
. PMID 9482895.
Further reading
- Lehner B (2007). "Modelling genotype-phenotype relationships and human disease with genetic interaction networks". J Exp Biol. 210 (9): 1559–66. doi:10.1242/jeb.002311. PMID 17449820.
- Genetic Science Learning Center. "Homeotic Genes and Body Patterns." Learn.Genetics 6 April 2016 <http://learn.genetics.utah.edu/content/variation/hoxgenes/>
- Guo, H.-S., et al. "Evolution Of The KCS Gene Family In Plants: The History Of Gene Duplication, Sub/Neofunctionalization And Redundancy.[1]" Molecular Genetics And Genomics (2015): 14p. Scopus®.
- Nowak Ma, Boerlijst Mc, Cooke J, Smith Jm. "Evolution of genetic redundancy". Nature 388, 167-171. (10 July 1997).
- ↑ Guo, Hai-Song; Zhang, Yan-Mei; Sun, Xiao-Qin; Li, Mi-Mi; Hang, Yue-Yu; Xue, Jia-Yu (2015-11-12). "Evolution of the KCS gene family in plants: the history of gene duplication, sub/neofunctionalization and redundancy". Molecular Genetics and Genomics: 1–14. doi:10.1007/s00438-015-1142-3. ISSN 1617-4615.