Cryo-electron tomography
Electron cryo-tomography (ECT, also called cryo-electron tomography, cryo-ET or CET) is an imaging technique used to produce high-resolution (~4 nm) three-dimensional views of samples, typically biological macromolecules and cells.[1] ECT is a specialized application of transmission electron microscopy (TEM) in which samples are imaged as they are tilted, resulting in a series of 2D images that can be combined to produce a 3D reconstruction, similar to a CT scan of the human body. In contrast to other electron tomography techniques, samples are immobilized in non-crystalline (“vitreous”) ice and imaged under cryogenic conditions (< -150 °C), allowing them to be imaged without dehydration or chemical fixation, which could otherwise disrupt or distort biological structures.[2]
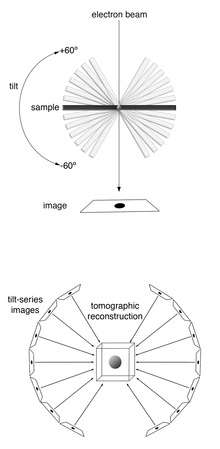
Description of technique
In electron microscopy (EM), samples are imaged in an ultra-high vacuum. Such a vacuum is incompatible with biological samples such as cells; the water would boil off, and the difference in pressure would explode the cell. In room-temperature EM techniques, samples are therefore prepared by fixation and dehydration. Another approach to stabilize biological samples, however, is to freeze them (cryo-electron microscopy). As in other cryo-electron microscopy techniques, samples for ECT (typically small cells (e.g. Bacteria or Archaea) or viruses) are prepared in standard aqueous media and applied to an EM grid. The grid is then plunged into a cryogen (typically ethane) so efficient that water molecules do not have time to rearrange into a crystalline lattice.[2] The resulting water state is called “vitreous ice” and preserves native cellular structures, such as lipid membranes, that would normally be destroyed by freezing. Plunge-frozen samples are subsequently stored and imaged at liquid-nitrogen temperatures so that the water never warms enough to crystallize.
Samples are imaged in a transmission electron microscope (TEM). As in other electron tomography techniques, the sample is tilted to different angles relative to the electron beam (typically every 1 or 2 degrees from about -60º degrees to +60º), and an image is acquired at each angle. This tilt-series of images can then be computationally reconstructed into a three-dimensional view of the object of interest.[3] This is called a tomogram, or tomographic reconstruction.
Applications
In transmission electron microscopy (TEM), because electrons interact strongly with matter, resolution is limited by the thickness of the sample. Therefore, for ECT, samples should be less than ~500 nm thick to achieve “macromolecular” resolution (~4 nm). For this reason, most ECT studies have focused on purified macromolecular complexes, viruses, or small cells such as those of many species of Bacteria and Archaea.[1]
Larger cells, and even tissues, can be prepared for ECT by thinning, either by cryo-sectioning or by focused ion beam (FIB)-milling. In cryo-sectioning, frozen blocks of cells or tissue are sectioned into thin samples with a cryo-microtome.[4] In FIB-milling, plunge-frozen samples are exposed to a focused beam of ions, typically gallium, that precisely whittle away material from the top and bottom of a sample, leaving a thin lamella suitable for ECT imaging.[5]
The strong interaction of electrons with matter also results in an anisotropic resolution effect. As the sample is tilted during imaging, the electron beam interacts with a relatively greater cross-sectional area at higher tilt angles. In practice, tilt angles greater than approximately ±60-70º do not yield much information and are therefore not used. This results in a “missing wedge” of information in the final tomogram that decreases resolution parallel to the electron beam.[3]
For structures that are present in multiple copies in one or multiple tomograms, higher resolution (even ≤1 nm) can be obtained by subtomogram averaging.[6][7] Similar to single particle analysis, subtomogram averaging computationally combines images of identical objects to increase the signal-to-noise ratio.
A major obstacle in ECT is identifying structures of interest within complicated cellular environments. One solution is to apply correlated cryo-fluorescence light microscopy,[8] and even super-resolution light microscopy (e.g. cryo-PALM[9]), and ECT. In these techniques, a sample containing a fluorescently-tagged protein of interest is plunge-frozen and first imaged in a light microscope equipped with a special stage to allow the sample to be kept at sub-crystallization temperatures (<150 °C). The location of the fluorescent signal is identified and the sample is transferred to the cryo-TEM, where the same location is then imaged at high resolution by ECT.
See also
References
- 1 2 Gan, Lu; Jensen, Grant J. (2012-02-01). "Electron tomography of cells". Quarterly Reviews of Biophysics. 45 (1): 27–56. doi:10.1017/S0033583511000102. ISSN 1469-8994. PMID 22082691.
- 1 2 Dubochet, J.; Adrian, M.; Chang, J. J.; Homo, J. C.; Lepault, J.; McDowall, A. W.; Schultz, P. (1988-05-01). "Cryo-electron microscopy of vitrified specimens". Quarterly Reviews of Biophysics. 21 (2): 129–228. doi:10.1017/s0033583500004297. ISSN 0033-5835. PMID 3043536.
- 1 2 Lučič, Vladan; Rigort, Alexander; Baumeister, Wolfgang (2013-08-05). "Cryo-electron tomography: the challenge of doing structural biology in situ". The Journal of Cell Biology. 202 (3): 407–419. doi:10.1083/jcb.201304193. ISSN 1540-8140. PMC 3734081
 . PMID 23918936.
. PMID 23918936. - ↑ Al-Amoudi, Ashraf; Chang, Jiin-Ju; Leforestier, Amélie; McDowall, Alasdair; Salamin, Laurée Michel; Norlén, Lars P. O.; Richter, Karsten; Blanc, Nathalie Sartori; Studer, Daniel (2004-09-15). "Cryo-electron microscopy of vitreous sections". The EMBO journal. 23 (18): 3583–3588. doi:10.1038/sj.emboj.7600366. ISSN 0261-4189. PMC 517607
 . PMID 15318169.
. PMID 15318169. - ↑ Villa, Elizabeth; Schaffer, Miroslava; Plitzko, Jürgen M.; Baumeister, Wolfgang (2013-10-01). "Opening windows into the cell: focused-ion-beam milling for cryo-electron tomography". Current Opinion in Structural Biology. 23 (5): 771–777. doi:10.1016/j.sbi.2013.08.006. ISSN 1879-033X. PMID 24090931.
- ↑ Briggs, John A. G. (2013-04-01). "Structural biology in situ--the potential of subtomogram averaging". Current Opinion in Structural Biology. 23 (2): 261–267. doi:10.1016/j.sbi.2013.02.003. ISSN 1879-033X. PMID 23466038.
- ↑ Schur, Florian K. M.; Dick, Robert A.; Hagen, Wim J. H.; Vogt, Volker M.; Briggs, John A. G. (2015-10-15). "The Structure of Immature Virus-Like Rous Sarcoma Virus Gag Particles Reveals a Structural Role for the p10 Domain in Assembly". Journal of Virology. 89 (20): 10294–10302. doi:10.1128/JVI.01502-15. ISSN 1098-5514. PMC 4580193
 . PMID 26223638.
. PMID 26223638. - ↑ Zhang, Peijun (2013-10-01). "Correlative cryo-electron tomography and optical microscopy of cells". Current Opinion in Structural Biology. 23 (5): 763–770. doi:10.1016/j.sbi.2013.07.017. ISSN 1879-033X. PMC 3812453
 . PMID 23962486.
. PMID 23962486. - ↑ Chang, Yi-Wei; Chen, Songye; Tocheva, Elitza I.; Treuner-Lange, Anke; Löbach, Stephanie; Søgaard-Andersen, Lotte; Jensen, Grant J. (2014-07-01). "Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography". Nature Methods. 11 (7): 737–739. doi:10.1038/nmeth.2961. ISSN 1548-7105. PMC 4081473
 . PMID 24813625.
. PMID 24813625.