Complex systems biology
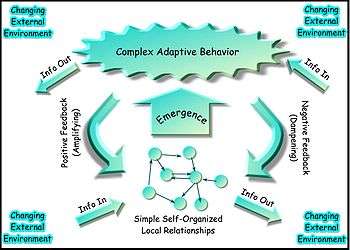
Complex systems biology (CSB) is a branch or subfield of mathematical and theoretical biology concerned with complexity of both structure and function in biological organisms, as well as the emergence and evolution of organisms and species, with emphasis being placed on the complex interactions of, and within, bionetworks,[1] and on the fundamental relations and relational patterns that are essential to life.[2][3][4][5][6] CSB is thus a field of theoretical sciences aimed at discovering and modeling the relational patterns essential to life that has only a partial overlap with complex systems theory,[7] and also with the systems approach to biology called systems biology; this is because the latter is restricted primarily to simplified models of biological organization and organisms, as well as to only a general consideration of philosophical or semantic questions related to complexity in biology. Moreover, a wide range of abstract theoretical complex systems are studied as a field of applied mathematics, with or without relevance to biology, chemistry or physics.
Complexity of organisms and biosphere
A complete definition of complexity for individual organisms, species, ecosystems, biological evolution and the biosphere has eluded researchers, and still is an ongoing issue.[3][8]
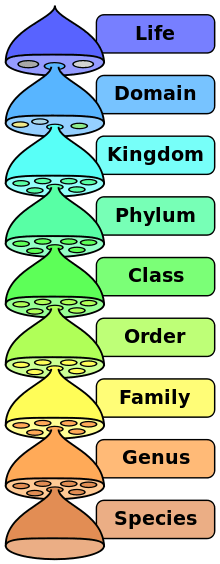
Most complex system models are often formulated in terms of concepts drawn from statistical physics, information theory and non-linear dynamics; however, such approaches are not focused on, or do not include, the conceptual part of complexity related to organization and topological attributes or algebraic topology, such as network connectivity of genomes, interactomes and biological organisms that are very important.[6][9][10] Recently, the two complementary approaches based both on information theory, network topology/abstract graph theory concepts are being combined for example in the fields of neuroscience and human cognition.[7][11] It is generally agreed that there is a hierarchy of complexity levels of organization that should be considered as distinct from that of the levels of reality in ontology.[7][12][13] The hierarchy of complexity levels of organization in the biosphere is also recognized in modern classifications of taxonomic ranks, such as: biological domain and biosphere, biological kingdom, Phylum, biological class, order, family, genus and species. Because of their dynamic and composition variability, intrinsic "fuzziness", autopoietic attributes, ability to self-reproduce, and so on, organisms do not fit into the 'standard' definition of general systems, and they are therefore 'super-complex' in both their function and structure; organisms can be thus be defined in CSB only as 'meta-systems' of simpler dynamic systems[7][14] Such a meta-system definition of organisms, species, 'ecosystems', and so on, is not equivalent to the definition of a system of systems as in Autopoietic Systems Theory,;[15] it also differs from the definition proposed for example by K.D. Palmer in meta-system engineering,[16] organisms being quite different from machines and automata with fixed input-output transition functions, or a continuous dynamical system with fixed phase space,[17] contrary to the Cartesian philosophical thinking; thus, organisms cannot be defined merely in terms of a quintuple A of (states, startup state, input and output sets/alphabet, transition function),[18] although 'non-deterministic automata', as well as 'fuzzy automata' have also been defined. Tessellation or cellular automata provide however an intuitive, visual/computational insight into the lower levels of complexity, and have therefore become an increasingly popular, discrete model studied in computability theory, applied mathematics, physics, computer science, theoretical biology/systems biology, cancer simulations and microstructure modeling. Evolving cellular automata using genetic algorithms[19][20][21] is also an emerging field attempting to bridge the gap between the tessellation automata and the higher level complexity approaches in CSB.
Topics in complex systems biology

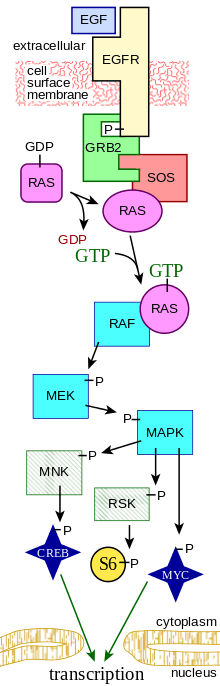
The following is only a partial list of topics covered in complex systems biology:
- Organisms and species relations and evolution
- Interactions among Species
- Evolution theories and population genetics
- Population genetics models
- Epigenetics
- Molecular evolution theories
- Quantum biocomputation
- Quantum genetics[22]
- Relational biology[4][23][24][25]
- Self-reproduction[26] (also called self-replication in a more general context)
- Computational gene models
- Evolutionary developmental biology
- Autopoiesis
- Protein folding
- Telomerase conformations and functions in vivo
- Epigenetics
- Interactomics[27][28]
- Cell signaling
- Signal transduction networks
- Complex neural nets
- Genetic networks
- Morphogenesis
- Digital morphogenesis
- Complex adaptive systems
- Topological models of morphogenesis
- Population dynamics of fisheries
- Epidemiology
- Theoretical ecology
- Immune system
Related journals
- Acta Biotheoretica
- Bioinformatics
- Biological Theory
- BioSystems
- Bulletin of Mathematical Biology
- Ecological Modelling
- Journal of Mathematical Biology
- Journal of Theoretical Biology
- Mathematical Biosciences
- Medical Hypotheses
- Theoretical and Applied Genetics
- Theoretical Biology and Medical Modelling
- Theoretical Population Biology
- Theory in Biosciences (formerly: Biologisches Zentralblatt)
CBS societies and institutes
- Society for Mathematical Biology
- ESMTB: European Society for Mathematical and Theoretical Biology
- Division of Mathematical Biology at NIMR
- The Israeli Society for Theoretical and Mathematical Biology
- Société Francophone de Biologie Théorique
- International Society for Biosemiotic Studies
See also
Biographies
Further reading
- A general list of Theoretical biology/Mathematical biology references, including an updated list of actively contributing authors.
- A list of references for applications of category theory in relational biology.
- An updated list of publications of theoretical biologist Robert Rosen
- Theory of Biological Anthropology (Documents No. 9 and 10 in English)
- Drawing the Line Between Theoretical and Basic Biology, a forum article by Isidro A. T. Savillo
- Synthesis and Analysis of a Biological System, by Hiroyuki Kurata, 1999.
Notes
- ↑ Sprites, P; Glymour, C; Scheines, R (2000). Causation, Prediction, and Search: Adaptive Computation and Machine Learning (2nd ed.). MIT Press.
- ↑ Donald Snooks, Graeme, "A general theory of complex living systems: Exploring the demand side of dynamics", Complexity, vol. 13, no. 6, July/August 2008.
- 1 2 Bonner, J. T. 1988. The Evolution of Complexity by Means of Natural Selection. Princeton: Princeton University Press.
- 1 2 Rosen, R. (1958a). "A Relational Theory of Biological Systems". Bulletin of Mathematical Biophysics. 20: 245–260. doi:10.1007/bf02478302.
- ↑ Baianu, I. C. (2006). "Robert Rosen's Work and Complex Systems Biology". Axiomathes. 16 (1-2): 25–34. doi:10.1007/s10516-005-4204-z.
- 1 2 Rosen, R. (1958b). "The Representation of Biological Systems from the Standpoint of the Theory of Categories". Bulletin of Mathematical Biophysics. 20: 317–341. doi:10.1007/bf02477890.
- 1 2 3 4 Baianu, I. C.; Brown, R.; Glazebrook, J. F. (2007). "Categorical Ontology of Complex Spacetime Structures: The Emergence of Life and Human Consciousness". Axiomathes. 17: 223–352. doi:10.1007/s10516-007-9011-2.
- ↑ Heylighen, Francis (2008). "Complexity and Self-Organization". In Bates, Marcia J.; Maack, Mary Niles. Encyclopedia of Library and Information Sciences. CRC. ISBN 978-0-8493-9712-7
- ↑ ^ Heylighen, Francis (2008). "Complexity and Self-Organization". In Bates, Marcia J.; Maack, Mary Niles. Encyclopedia of Library and Information Sciences. CRC. ISBN 978-0-8493-9712-7
- ↑ "abstract relational biology (ARB)". PlanetPhysics. Retrieved 2010-03-17.
- ↑ http://hdl.handle.net/10101/npre.2011.6115.1 Wallace, Rodrick. When Spandrels Become Arches: Neural crosstalk and the evolution of consciousness. Available from Nature Precedings (2011)
- ↑ Poli R (2001a). "The Basic Problem of the Theory of Levels of Reality". Axiomathes. 12 (3-4): 261–283.
- ↑ Poli R (1998). "Levels". Axiomathes. 9 (1-2): 197–211. doi:10.1007/bf02681712.
- ↑ Metasystem Transition Theory, Valentin Turchin, Cliff Joslyn, 1993-1997
- ↑ Reflexive Autopoietic Systems Theory
- ↑ Meta-system Engineering, Kent D. Palmer, 1996
- ↑ Hoff, M.A., Roggia, K.G., Menezes, P.B.:(2004). Composition of Transformations: A Framework for Systems with Dynamic Topology. International Journal of Computing Anticipatory Systems 14:259–270
- ↑ John E. Hopcroft, Rajeev Motwani, Jeffrey D. Ullman.2000. Introduction to Automata Theory, Languages, and Computation (2nd Edition)Pearson Education. ISBN 0-201-44124-1
- ↑ The Evolutionary Design of Collective Computation in Cellular Automata, James P. Crutchfeld, Melanie Mitchell, Rajarshi Das (In J. P. Crutchfield and P. K. Schuster (editors), Evolutionary Dynamics|Exploring the Interplay of Selection, Neutrality, Accident, and Function. New York: Oxford University Press, 2002.)
- ↑ Evolving Cellular Automata with Genetic Algorithms: A Review of Recent Work, Melanie Mitchell, James P. Crutchfeld, Rajarshi Das (In Proceedings of the First International Conference on Evolutionary Computation and Its Applications (EvCA'96). Moscow, Russia: Russian Academy of Sciences, 1996.)
- ↑ Peak, West; Messinger, Mott; Messinger, SM; Mott, KA (2004). "Evidence for complex, collective dynamics and emergent, distributed computation in plants". Proceedings of the National Academy of Sciences of the USA. 101 (4): 918–922. Bibcode:2004PNAS..101..918P. doi:10.1073/pnas.0307811100. PMC 327117
 . PMID 14732685.
. PMID 14732685. - ↑ Rosen, R. 1960. (1960). "A quantum-theoretic approach to genetic problems". Bulletin of Mathematical Biophysics. 22 (3): 227–255. doi:10.1007/BF02478347.
- ↑ Baianu, I. C.: 2006, (2006). "Robert Rosen's Work and Complex Systems Biology". Axiomathes. 16 (1–2): 25–34. doi:10.1007/s10516-005-4204-z.
- ↑ Rosen, R.: 1958b, (1958). "The Representation of Biological Systems from the Standpoint of the Theory of Categories". Bulletin of Mathematical Biophysics. 20 (4): 317–341. doi:10.1007/BF02477890.
- ↑ PlanetMath
- ↑ "PlanetMath". PlanetMath. Retrieved 2010-03-17.
- 1 2 Faith, JJ; et al. (2007). "Large-Scale Mapping and Validation of Escherichia coli Transcriptional Regulation from a Compendium of Expression Profiles". PLoS Biology. 5 (1): 54–66. doi:10.1371/journal.pbio.0050008. PMC 1764438
 . PMID 17214507.
. PMID 17214507. - 1 2 Hayete, B; Gardner, TS; Collins, JJ (2007). "Size matters: network inference tackles the genome scale". Molecular Systems Biology. 3 (1): 77. doi:10.1038/msb4100118. PMC 1828748
 . PMID 17299414.
. PMID 17299414. - ↑ "abstract relational biology (ARB)". PlanetPhysics. Archived from the original on January 7, 2016. Retrieved 2010-03-17.
- ↑ "Molecular Evolution and Protobiology | KLI Theory Lab". Theorylab.org. 2009-05-26. Retrieved 2010-03-17.
References cited
- Ahmed, E. (2006). "On modeling the immune system as a complex system". Theor. BioSci. 124: 413.
- Baianu, I. C., Computer Models and Automata Theory in Biology and Medicine., Monograph, Ch.11 in M. Witten (Editor), Mathematical Models in Medicine, vol. 7., Vol. 7: 1513-1577 (1987),Pergamon Press:New York, (updated by Hsiao Chen Lin in 2004 ISBN 0-08-036377-6
- Bonner, J. T. 1988. The Evolution of Complexity by Means of Natural Selection. Princeton: Princeton University Press.
- Donald Snooks, Graeme, "A general theory of complex living systems: Exploring the demand side of dynamics", Complexity, vol. 13, no. 6, July/August 2008.
- Drazin, P.G., Nonlinear systems. C.U.P., 1992. ISBN 0-521-40668-4
- Edelstein-Keshet, L., Mathematical Models in Biology. SIAM, 2004. ISBN 0-07-554950-6
- Forgacs, G.; S. A. Newman, Biological Physics of the Developing Embryo. C.U.P., 2005. ISBN 0-521-78337-2
- Israel G (1988). "On the contribution of Volterra and Lotka to the development of modern biomathematics". History and Philosophy of the Life Sciences. 10 (1): 37–49. PMID 3045853.
- Israel, G., 2005, "Book on mathematical biology" in Grattan-Guinness, I., ed., Landmark Writings in Western Mathematics. Elsevier: 936-44.
- Jordan, D.W.; Smith, P., Nonlinear ordinary differential equations, 2nd ed. O.U.P., 1987. ISBN 0-19-856562-3
- Kampen, N.G. van. Stochastic Processes in Physics and Chemistry, North Holland., 3rd ed. 2001, ISBN 0-444-89349-0
- Murray, J.D., Mathematical Biology. Springer-Verlag, 3rd ed. in 2 vols.: Mathematical Biology: I. An Introduction, 2002 ISBN 0-387-95223-3; Mathematical Biology: II. Spatial Models and Biomedical Applications, 2003 ISBN 0-387-95228-4.
- Nicolas Rashevsky. (1938)., Mathematical Biophysics. Chicago: University of Chicago Press.
- Preziosi, L., Cancer Modelling and Simulation. Chapman Hall/CRC Press, 2003. ISBN 1-58488-361-8.
- Renshaw, E., Modelling biological populations in space and time. C.U.P., 1991. ISBN 0-521-44855-7
- Rosen, Robert.1991, Life Itself: A Comprehensive Inquiry into the Nature, Origin, and Fabrication of Life, Columbia University Press, published posthumously:
- Rosen, Robert .1970. Dynamical system theory in biology. New York, Wiley-Interscience. ISBN 0-471-73550-7
- Rosen, Robert. 2000, Essays on Life Itself, Columbia University Press.
- Rosen, Robert. 2003, "Anticipatory Systems; Philosophical, Mathematical, and Methodolical Foundations", Rosen Enterprises publs.
- Rubinow, S.I., Introduction to mathematical biology. John Wiley, 1975. ISBN 0-471-74446-8
- Scudo FM (March 1971). "Vito Volterra and theoretical ecology". Theoretical Population Biology. 2 (1): 1–23. doi:10.1016/0040-5809(71)90002-5. PMID 4950157.
- Segel, L.A., Modeling dynamic phenomena in molecular and cellular biology. C.U.P., 1984. ISBN 0-521-27477-X.
- Strogatz, S.H., Nonlinear dynamics and Chaos: Applications to Physics, Biology, Chemistry, and Engineering. Perseus, 2001, ISBN 0-7382-0453-6
- Thompson, D'Arcy W., 1992. On Growth and Form. Dover reprint of 1942, 2nd ed. (1st ed., 1917). ISBN 0-486-67135-6
External links
- Center for Complex Systems and Brain Sciences at Florida Atlantic
- Santa Fe Institute
- Bulletin of Mathematical Biology
- European Society for Mathematical and Theoretical Biology
- Journal of Mathematical Biology
- Biomathematics Research Centre at University of Canterbury
- Centre for Mathematical Biology at Oxford University
- Mathematical Biology at the National Institute for Medical Research
- Institute for Medical BioMathematics
- Mathematical Biology Systems of Differential Equations from EqWorld: The World of Mathematical Equations
- Systems Biology Workbench - a set of tools for modelling biochemical networks
- The Collection of Biostatistics Research Archive
- Statistical Applications in Genetics and Molecular Biology
- The International Journal of Biostatistics
- Theoretical Modeling of Cellular Physiology at Ecole Normale Superieure, Paris
- Theoretical and mathematical biology website
- Complexity Discussion Group
- UCLA Biocybernetics Laboratory
- TUCS Computational Biomodelling Laboratory
- Nagoya University Division of Biomodeling
- Technische Universiteit Biomodeling and Informatics
- New England Complex Systems Institute
- Northwestern Institute on Complex Systems (NICO)
- Complexity Digest
- Centro de Ciencias de la Complejidad, UNAM
- Complexity Complex at the University of Warwick
- Southampton Institute for Complex Systems Simulation
- Center for the Study of Complex Systems at the University of Michigan
- ARC Centre for Complex Systems, Australia
- (European) Complex Systems Society
- (Australian) Complex systems research network.
- Complex Systems Modeling based on Luis M. Rocha, 1999.
- CRM Complex systems research group