Natural competence
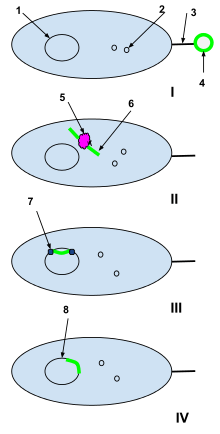
1-Bacterial cell DNA
2-Bacterial cell plasmids
3-Sex pili
4-Plasmid of foreign DNA from a dead cell
5-Bacterial cell restriction enzyme
6-Unwound foreign plasmid
7-DNA ligase
I: A plasmid of foreign DNA from a dead cell is intercepted by the sex pili of a naturally competent bacterial cell.
II: The foreign plasmid is transduced through the sex pili into the bacterial cell, where it is processed by bacterial cell restriction enzymes. The restriction enzymes break the foreign plasmid into a strand of nucleotides that can be added to the bacterial DNA.
III: DNA ligase integrates the foreign nucleotides into the bacterial cell DNA.
IV: Recombination is complete and the foreign DNA has integrated into the original bacterial cell’s DNA and will continue to be a part of it when the bacterial cell replicates next.
In microbiology, genetics, cell biology, and molecular biology, competence is the ability of a cell to take up extracellular ("naked") DNA from its environment in the process called transformation. Competence may be differentiated between natural competence, a genetically specified ability of bacteria which is thought to occur under natural conditions as well as in the laboratory, and induced or artificial competence, which arises when cells in laboratory cultures are treated to make them transiently permeable to DNA. This article primarily deals with natural competence in bacteria, although information about artificial competence is also provided.
History
Natural competence was discovered by Frederick Griffith in 1928, when he showed that a preparation of killed cells of a pathogenic bacterium contained something that could transform related non-pathogenic cells into the pathogenic type. In 1944 Oswald Avery, Colin MacLeod, and Maclyn McCarty demonstrated that this 'transforming factor' was pure DNA[1] . This was the first compelling evidence that DNA carries the genetic information of the cell.
Since then, natural competence has been studied in a number of different bacteria, particularly Bacillus subtilis, Streptococcus pneumoniae (Griffith's "pneumococcus"), Neisseria gonorrhoeae and Haemophilus influenzae. Areas of active research include the mechanisms of DNA transport, the regulation of competence in different bacteria, and the evolutionary function of competence.
Mechanisms of DNA uptake
In the natural world DNA usually becomes available by death and lysis of other cells, but in the laboratory it is provided by the researcher, often as a genetically engineered fragment or plasmid. During uptake, DNA is transported across the cell membrane(s), and the cell wall if one is present. Once the DNA is inside the cell it may be degraded to nucleotides, which are reused for DNA replication and other metabolic functions. Alternatively it may be recombined into the cell’s genome by its DNA repair enzymes. If this recombination changes the cell’s genotype the cell is said to have been transformed. Artificial competence and transformation are used as research tools in many organisms (see Transformation (genetics)).[2]
In almost all naturally competent bacteria components of extracellular filaments called type 4 pili (a type of fimbria) bind extracellular double stranded DNA. The DNA is then translocated across the membrane (or membranes for gram negative bacteria) through multi-component protein complexes driven by the degradation of one strand of the DNA. Single stranded DNA in the cell is bound by a well-conserved protein, DprA, which loads the DNA onto RecA, which mediates homologous recombination through the classic DNA repair pathway.[3]
Regulation of competence
In laboratory cultures, natural competence is usually tightly regulated and often triggered by nutritional shortages or adverse conditions. However the specific inducing signals and regulatory machinery are much more variable than the uptake machinery, and little is known about the regulation of competence in the natural environments of these bacteria.[4] Transcription factors have been discovered which regulate competence; an example is sxy (also known as tfoX) which has been found to be regulated in turn by a 5' non-coding RNA element.[5] In bacteria capable of forming spores, conditions inducing sporulation often overlap with those inducing competence. Thus cultures or colonies containing sporulating cells often also contain competent cells. Recent research by Süel et al. has identified an excitable core module of genes which can explain entry into and exit from competence when cellular noise is taken into account.[6]
Most competent bacteria are thought to take up all DNA molecules with roughly equal efficiencies, but bacteria in the families Neisseriaceae and Pasteurellaceae preferentially take up DNA fragments containing short DNA sequences, termed DNA uptake sequence (DUS and USS respectively), that are very frequent in their own genomes. Neisserial genomes contain thousands of copies of the preferred sequence GCCGTCTGAA, and Pasteurellacean genomes contain either AAGTGCGGT or ACAAGCGGT.[2][7]
Evolutionary functions and consequences of competence
Most proposals made for the primary evolutionary function of natural competence as a part of natural bacterial transformation fall into three categories: (1) the selective advantage of genetic diversity; (2) DNA uptake as a source of nucleotides (DNA as “food”); and (3) the selective advantage of a new strand of DNA to promote homologous recombinational repair of damaged DNA (DNA repair). A secondary suggestion has also been made, noting the occasional advantage of horizontal gene transfer.
Hypothesis of genetic diversity
Arguments to support genetic diversity as the primary evolutionary function of sex (including bacterial transformation) are given by Barton and Charleworth .[8] and by Otto and Gerstein.[9] However, the theoretical difficulties associated with the evolution of sex suggest that sex for genetic diversity is problematic. Specifically with respect to bacterial transformation, competence requires the high cost of a global protein synthesis switch, with, for example, more than 16 genes that are switched on only during competence of Strepococcus pneumoniae.[10] However, since bacteria tend to grow in clones, the DNA available for transformation would generally have the same genotype as that of the recipient cells. Thus, there is always a high cost in protein expression without, in general, an increase in diversity. Other differences between competence and sex have been considered in models of the evolution of genes causing competence; these models found that competence's postulated recombinational benefits were even more elusive than those of sex.[11]
Hypothesis of DNA as food
The second hypothesis, DNA as food, relies on the fact that cells that take up DNA inevitably acquire the nucleotides the DNA consists of, and, because nucleotides are needed for DNA and RNA synthesis and are expensive to synthesize, these may make a significant contribution to the cell's energy budget.[12] Some naturally competent bacteria also secrete nucleases into their surroundings, and all bacteria can take up the free nucleotides these nucleases generate from environmental DNA.[13] The energetics of DNA uptake are not understood in any system, so it is difficult to compare the efficiency of nuclease secretion to that of DNA uptake and internal degradation. In principle the cost of nuclease production and the uncertainty of nucleotide recovery must be balanced against the energy needed to synthesize the uptake machinery and to pull DNA in. Other important factors are the likelihoods that nucleases and competent cells will encounter DNA molecules, the relative inefficiencies of nucleotide uptake from the environment and from the periplasm (where one strand is degraded by competent cells), and the advantage of producing ready-to-use nucleotide monophosphates from the other strand in the cytoplasm. Another complicating factor is the self-bias of the DNA uptake systems of species in the family Pasteurellaceae and the genus Neisseria, which could reflect either selection for recombination or for mechanistically efficient uptake.[14][15]
Hypothesis of repair of DNA damage
In bacteria, the problem of DNA damage is most pronounced during periods of stress, particularly oxidative stress, that occur during crowding or starvation conditions. Under such conditions there is often only a single chromosome present. The finding that some bacteria induce competence under such stress conditions, supports the third hypothesis, that transformation exists to permit DNA repair. In experimental tests, bacterial cells exposed to agents damaging their DNA, and then undergoing transformation, survived better than cells exposed to DNA damage that did not undergo transformation (Hoelzer and Michod, 1991).[16] In addition, competence to undergo transformation is often inducible by known DNA damaging agents (reviewed by Michod et al., 2008 and Bernstein et al., 2012).[17][18] Thus, a strong short-term selective advantage for natural competence and transformation would be its ability to promote homologous recombinational DNA repair under conditions of stress. Such stress conditions might be incurred during bacterial infection of a susceptible host. Consistent with this idea, Li et al. [19] reported that, among different highly transformable S. pneumoniae isolates, nasal colonization fitness and virulence (lung infectivity) depends on an intact competence system.
A counter argument was made based on the 1993 report of Redfield who found that single-stranded and double-stranded damage to chromosomal DNA did not induce or enhance competence or transformation in B. subtilis or H. influenzae, suggesting that selection for repair has played little or no role in the evolution of competence in these species[20]
However more recent evidence indicates that competence for transformation is, indeed, specifically induced by DNA damaging conditions. For instance, Claverys et al. in 2006[21] showed that the DNA damaging agents mitomycin C (a DNA cross-linking agent) and fluoroquinolone (a topoisomerase inhibitor that causes double-strand breaks) induce transformation in Streptococcus pneumoniae. In addition, Engelmoer and Rozen[22] in 2011 demonstrated that in S. pneumoniae transformation protects against the bactericidal effect of mitomycin C. Induction of competence further protected against the antibiotics kanomycin and streptomycin.[21][22] Although these aminoglycoside antibiotics were previously regarded as non-DNA damaging, recent studies in 2012 of Foti et al.[23] showed that a substantial portion of their bactericidal activity results from release of the hydroxyl radical and induction of DNA damages, including double-strand breaks.
Dorer et al.,[24] in 2010, showed that ciprofloxacin, which interacts with DNA gyrase and causes production of double-strand breaks, induces expression of competence genes in Helicobacter pylori, leading to increased transformation. In 2011 studies of Legionella pneumophila, Charpentier et al.[25] tested 64 toxic molecules to determine which ones induce competence. Only six of these molecules, all DNA damaging agents, strongly induced competence. These molecules were norfloxacin, ofloxacin and nalidixic acid (inhibitors of DNA gyrase that produce double strand breaks[26]), mitomycin C (which produces inter-strand cross-links), bicyclomycin (causes single- and double-strand breaks[27]), and hydroxyurea (causes oxidation of DNA bases[28]). Charpentier et al.[25] also showed that UV irradiation induces competence in L. pneumophila and further suggested that competence for transformation evolved as a response to DNA damage.
Horizontal gene transfer
A long-term advantage may occasionally be conferred by occasional instances of horizontal gene transfer also called lateral gene transfer, (which might result from non-homologous recombination after competence is induced), that could provide for antibiotic resistance or other advantages.
Regardless of the nature of selection for competence, the composite nature of bacterial genomes provides abundant evidence that the horizontal gene transfer caused by competence contributes to the genetic diversity that makes evolution possible.
References
- ↑ Avery OT, Macleod CM, McCarty M (1944). "STUDIES ON THE CHEMICAL NATURE OF THE SUBSTANCE INDUCING TRANSFORMATION OF PNEUMOCOCCAL TYPES : INDUCTION OF TRANSFORMATION BY A DESOXYRIBONUCLEIC ACID FRACTION ISOLATED FROM PNEUMOCOCCUS TYPE III". J. Exp. Med. 79 (2): 137–58. PMC 2135445
 . PMID 19871359.
. PMID 19871359. - 1 2 Chen I, Dubnau D (2004). "DNA uptake during bacterial transformation". Nat. Rev. Microbiol. 2 (3): 241–9. doi:10.1038/nrmicro844. PMID 15083159.
- ↑ Johnston C, Martin B, Fichant G, Polard P, Claverys J (2014). "Bacterial transformation: distribution, shared mechanisms and divergent control". Nat. Rev. Microbiol. 12: 181–96. doi:10.1038/nrmicro3199.
- ↑ Solomon JM, Grossman AD (1996). "Who's competent and when: regulation of natural genetic competence in bacteria". Trends Genet. 12 (4): 150–5. doi:10.1016/0168-9525(96)10014-7. PMID 8901420.
- ↑ Redfield RJ (September 1991). "sxy-1, a Haemophilus influenzae mutation causing greatly enhanced spontaneous competence". J. Bacteriol. 173 (18): 5612–8. PMC 208288
 . PMID 1653215.
. PMID 1653215. - ↑ Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB (2006). "An excitable gene regulatory circuit induces transient cellular differentiation". Nature. 440 (7083): 545–50. doi:10.1038/nature04588. PMID 16554821.
- ↑ Findlay, WA; Redfield, RJ (2009). "Coevolution of DNA uptake sequences and bacterial proteomes". Genome biology and evolution. 1: 45–55. doi:10.1093/gbe/evp005. PMC 2817400
 . PMID 20333176.
. PMID 20333176. - ↑ Barton NH, Charlesworth B (1998). "Why sex and recombination?". Science. 281 (5385): 1986–1990. doi:10.1126/science.281.5385.1986. PMID 9748151.
- ↑ Otto SP, Gerstein AC (Aug 2006). "Why have sex? The population genetics of sex and recombination". Biochem Soc Trans. 34 (Pt 4): 519–522. doi:10.1042/BST0340519. PMID 16856849.
- ↑ Peterson S, Cline RT, Tettelin H, Sharov V, Morrison DA (Nov 2000). "Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays". J Bacteriol. 182 (21): 6192–6202. doi:10.1128/JB.182.21.6192-6202.2000. PMC 94756
 . PMID 11029442.
. PMID 11029442. - ↑ Redfield R (1988). "Is sex with dead cells ever better than no sex at all?". Genetics. 119 (1): 213–21. PMC 1203342
 . PMID 3396864.
. PMID 3396864. - ↑ Redfield RJ (2001). "Do bacteria have sex?". Nat. Rev. Genet. 2 (8): 634–9. doi:10.1038/35084593. PMID 11483988.
- ↑ Dubnau D (1999). "DNA uptake in bacteria". Annu Rev Microbiol. 53 (1): 217–44. doi:10.1146/annurev.micro.53.1.217. PMID 10547691.
- ↑ Maughan H (2010). "Bacterial DNA uptake sequences can accumulate by molecular drive alone". Genetics. 186 (2): 613–27. doi:10.1534/genetics.110.119438. PMC 2954483
 . PMID 20628039.
. PMID 20628039. - ↑ Redfield R, Schrag M, Dead A (1997). "The evolution of bacterial transformation: sex with poor relations.". Genetics. 146 (1): 27–38. PMC 1207942
 . PMID 9135998.
. PMID 9135998. - ↑ Hoelzer MA, Michod RE (1991). "DNA repair and the evolution of transformation in Bacillus subtilis. III. Sex with damaged DNA". Genetics. 128 (2): 215–23. PMC 1204460
 . PMID 1906416.
. PMID 1906416. - ↑ Michod RE, Bernstein H, Nedelcu AM (2008). "Adaptive value of sex in microbial pathogens". Infect Genet Evol. 8 (3): 267–85. doi:10.1016/j.meegid.2008.01.002. PMID 18295550. http://www.hummingbirds.arizona.edu/Faculty/Michod/Downloads/IGE%20review%20sex.pdf
- ↑ Bernstein, Harris; Carol Bernstein; Richard E. Michod (2012). "Chapter 1 - DNA Repair as the Primary Adaptive Function of Sex in Bacteria and Eukaryotes". DNA Repair: New Research. NOVA Publishers. pp. 1–50. ISBN 978-1-62100-756-2. https://www.novapublishers.com/catalog/product_info.php?products_id=31918
- ↑ Li G, Liang Z, Wang X, Yang Y, Shao Z, Li M, Ma Y, Qu F, Morrison DA, Zhang JR (2016). "Addiction of Hypertransformable Pneumococcal Isolates to Natural Transformation for In Vivo Fitness and Virulence". Infect. Immun. 84 (6): 1887–901. doi:10.1128/IAI.00097-16. PMID 27068094.
- ↑ Redfield R (1993). "Evolution of natural transformation: testing the DNA repair hypothesis in Bacillus subtilis and Haemophilus influenzae". Genetics. 133 (4): 755–61. PMC 1205397
 . PMID 8462839.
. PMID 8462839. - 1 2 Claverys, JP; Prudhomme, M; Martin, B (2006). "Induction of competence regulons as a general response to stress in gram-positive bacteria". Annu Rev Microbiol. 60 (1): 451–475. doi:10.1146/annurev.micro.60.080805.142139. PMID 16771651.
- 1 2 Engelmoer, D J; Rozen, D E (2011). "Competence increases survival during stress in Streptococcus pneumoniae". Evolution. 65 (12): 3475–3485. doi:10.1111/j.1558-5646.2011.01402.x. PMID 22133219.
- ↑ Foti, JJ; Devadoss, B; Winkler, JA; Collins, JJ; Walker, GC (2012). "Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics". Science. 336 (6079): 315–319. doi:10.1126/science.1219192. PMC 3357493
 . PMID 22517853.
. PMID 22517853. - ↑ Dorer, MS; Fero, J; Salama, NR (2010). "DNA damage triggers genetic exchange in Helicobacter pylori". PLoS Pathog. 6 (7): e1001026. doi:10.1371/journal.ppat.1001026. PMC 2912397
 . PMID 20686662.
. PMID 20686662. - 1 2 Charpentier, X; Kay, E; Schneider, D; Shuman, HA (2011). "Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila". J Bacteriol. 193 (5): 1114–1121. doi:10.1128/JB.01146-10. PMC 3067580
 . PMID 21169481.
. PMID 21169481. - ↑ Albertini, S; Chételat, A A; Miller, B; Muster, W; Pujadas, E; Strobel, R; Gocke, E (1995). "Genotoxicity of 17 gyrase- and four mammalian topoisomerase II-poisons in prokaryotic and eukaryotic test systems". Mutagenesis. 10 (4): 343–351. doi:10.1093/mutage/10.4.343. PMID 7476271.
- ↑ Washburn, R S; Gottesman, M E (2011). "Transcription termination maintains chromosome integrity". Proc Natl Acad Sci U S A. 108 (2): 792–7. doi:10.1073/pnas.1009564108. PMC 3021005
 . PMID 21183718.
. PMID 21183718. - ↑ Sakano, K; Oikawa, S; Hasegawa, K; Kawanishi, S (2001). "Hydroxyurea induces site-specific DNA damage via formation of hydrogen peroxide and nitric oxide". Jpn J Cancer Res. 92 (11): 1166–1174. doi:10.1111/j.1349-7006.2001.tb02136.x. PMID 11714440.