Bottom-up proteomics
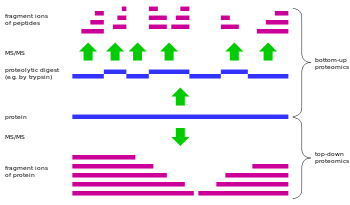
Bottom-up proteomics is a common method to identify proteins and characterize their amino acid sequences and post-translational modifications by proteolytic digestion of proteins prior to analysis by mass spectrometry.[1][2] The major alternative workflow used in high-throughput proteomics is called top-down proteomics and does not use proteolytic digestion.
In bottom-up proteomics, the proteins may first be purified by a method such as gel electrophoresis resulting in one or a few proteins in each proteolytic digest. Alternatively, the crude protein extract is digested directly, followed by one or more dimensions of separation of the peptides by liquid chromatography coupled to mass spectrometry, a technique known as shotgun proteomics.[3][4] By comparing the masses of the proteolytic peptides or their tandem mass spectra with those predicted from a sequence database or annotated peptide spectral in a peptide spectral library, peptides can be identified and multiple peptide identifications assembled into a protein identification.
Advantages
There is better front-end separation of peptides compared with proteins and higher sensitivity than the top-down method.[5]
Disadvantages
There is limited protein sequence coverage by identified peptides, loss of labile PTMs, and ambiguity of the origin for redundant peptide sequences.[5]
See also
References
- ↑ Aebersold R, Mann M (March 2003). "Mass spectrometry-based proteomics". Nature. 422 (6928): 198–207. doi:10.1038/nature01511. PMID 12634793.
- ↑ Chait BT (2006). "Chemistry. Mass spectrometry: bottom-up or top-down?". Science. 314 (5796): 65–6. doi:10.1126/science.1133987. PMID 17023639.
- ↑ Washburn MP, Wolters D, Yates JR (2001). "Large-scale analysis of the yeast proteome by multidimensional protein identification technology". Nat. Biotechnol. 19 (3): 242–247. doi:10.1038/85686. PMID 11231557.
- ↑ Wolters DA, Washburn MP, Yates JR (2001). "An automated multidimensional protein identification technology for shotgun proteomics". Anal. Chem. 73 (23): 5683–5690. doi:10.1021/ac010617e. PMID 11774908.
- 1 2 Yates JR, Ruse CI, Nakorchevsky A (2009). "Proteomics by Mass Spectrometry: Approaches, Advances, and Applications" (PDF). Annu. Rev. Biomed. Eng. 11: 49–79. doi:10.1146/annurev-bioeng-061008-124934. PMID 19400705.