Bioremediation of radioactive waste

Bioremediation of radioactive waste or bioremediation of radionuclides is an application of bioremediation based on the use of biological agents bacteria, plants and fungi (natural or genetically modified) to catalyze chemical reactions that allow the decontamination of sites affected by radionuclides.[2] These radioactive particles are by-products generated as a result of activities related to nuclear energy and constitute a pollution and a radiotoxicity problem (with serious health and ecological consequences) due to its unstable nature of ionizing radiation emissions.
The techniques of bioremediation of environmental areas as soil, water and sediments contaminated by radionuclides are diverse and currently being set up as an ecological and economic alternative to traditional procedures. Physico-chemical conventional strategies are based on the extraction of waste by excavating and drilling, with a subsequent long-range transport for their final confinement. These works and transport have often unacceptable estimated costs of operation that could exceed a trillion dollars in the US and 50 million pounds in the UK.[3]
The species involved in these processes have the ability to influence the properties of radionuclides such as solubility, bioavailability and mobility to accelerate its stabilization. Its action is largely influenced by electron donors and acceptors, nutrient medium, complexation of radioactive particles with the material and environmental factors. These are measures that can be performed on the source of contamination (in situ) or in controlled and limited facilities in order to follow the biological process more accurately and combine it with other systems (ex situ).[4][5]
Areas contaminated by radioactivity
Typology of radionuclides and polluting waste
The presence of radioactive waste in the environment may cause long-term effects due to the activity and half-life of the radionuclides, leading their impact to grow with time.[3] These particles exist in various oxidation states and are found as oxides, coprecipitates, or as organic or inorganic complexes, according to their origin and ways of liberation. Most commonly they are found in oxidized form, which makes them more soluble in water and thus more mobile.[5] Unlike organic contaminants, however, they cannot be destroyed and must be converted into a stable form or extracted from the environment.[6]
The sources of radioactivity are not exclusive of human activity. The natural radioactivity does not come from human sources: it covers up to ¾ of the total radioactivity in the world and has its origins in the interaction of terrestrial elements with high energy cosmic rays (cosmogenic radionuclides) or in the existing materials on Earth since its formation (primordial radionuclides). In this regard, there are differences in the levels of radioactivity throughout the Earth's crust. India and mountains like the Alps are among the areas with the highest level of natural radioactivity due to their composition of rocks and sand.[7]
The most frequent radionuclides in soils are naturally radium-226 (226Rn), radon-222 (222Rn), thorium-232 (232Th), uranium-238 (238U) and potassium-40 (40K). Potassium-40 (up to 88% of total activity), carbon-14 (14C), radium-226, uranium-238 and rubidium-87 (87Rb) are found in ocean waters. Moreover, in groundwater abound radius radioisotopes such as radium-226 and radium-228 (228Ra).[8][9] They are also habitual in building materials radionuclides of uranium, thorium and potassium (the latter common to wood).[9]
At the same time, anthropogenic radionuclides (caused by humans) are due to thermonuclear reactions resulting from explosions and nuclear weapons tests, discharges from nuclear facilities, accidents deriving from the reprocessing of commercial fuel, waste storage from these processes and to a lesser extent, nuclear medicine.[10] Some polluted sites by these radionuclides are the US DOE facilities (like Hanford Site), the Chernobyl and Fukushima exclusion zones and the affected area of Chelyabinsk Oblast due to the Kyshtym disaster.
In ocean waters, the presence of tritium (3H), cesium-137 (137Cs), strontium-90 (90Sr), plutonium-239 (239Pu) and plutonium-240 (240Pu) has significantly increased due to anthropogenic causes.[11][12] In soils, technetium-99 (99Tc), carbon-14, strontium-90, cobalt-60 (60Co), iodine-129 (129I), iodine-131 (131I), americium-241 (241Am), neptunium-237 (237Np) and various forms of radioactive plutonium and uranium are the most common radionuclides.[3][9][10]
| Frequency of occurrence of selected radionuclides at US DOE facilites | |||||
| Ground water | Soils/Sediments | ||||
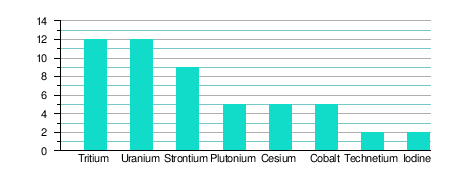 |
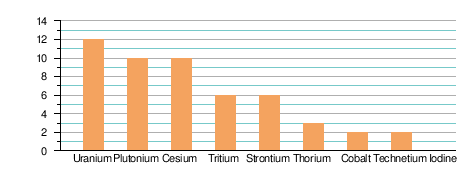 | ||||
| Source: United States Department of Energy, US Government (1992)[13] | |||||
The classification of radioactive waste established by the International Atomic Energy Agency (IAEA) distinguishes six levels according to equivalent dose, specific activity, heat released and half-life of the radionuclides:[14]
- Exempt waste (EW): Waste that meets the criteria for exclusion from regulatory control for radiation protection purposes.
- Very short lived waste (VSLW): Waste with very short half-lives (often used for research and medical purposes) that can be stored over a limited period of up to a few years and subsequently cleared from regulatory control.
- Very low level waste (VLLW): Waste like soil and rubble (with low levels of activity concentration) that may also contain other hazardous waste.
- Low level waste (LLW): Waste that is above clearance levels and requires robust isolation and containment for periods of up to a few hundred years and is suitable for disposal in engineered near surface facilities. LLW include short lived radionuclides at higher levels of activity concentration and also long lived radionuclides, but only at relatively low levels of activity concentration.
- Intermediate level waste (ILW): Waste with long lived radionuclides that requires a greater degree of containment and isolation at greater depths.
- High level waste (HLW): Waste with large amounts of long lived radionuclides that need to be stored in deep, stable geological formations usually several hundred metres or more below the surface.
Ecological and human health consequences
Radioactive contamination is a potential danger for living organisms and results in external hazards, concerning radiation sources outside the body, and internal dangers, as a result of the incorporation of radionuclides inside the body (often by inhalation of particles or ingestion of contaminated food).[15]
In humans, single doses from 0.25 Sv produce first anomalies in the amount of leukocytes. This effect is accentuated if the absorbed dose is between 0.5 and 2 Sv, in whose first damage, nausea and hair loss are suffered. The strip ranging between 2 and 5 Sv is considered the most serious and include bleeding, ulcers and risk of death; values exceeding 5 Sv involve immediate death.[15] If radiation, likewise, is received in small doses over long periods of time, the consequences can be equally severe. It is difficult to quantify the health effects for doses below 10 mSv, but it has been shown that there is a direct relationship between prolonged exposure and cancer risk (although there is not a very clear dose-response relationship to establish clear limits of exposure).[16]
The information available on the effect of natural background radiation with respect anthropogenic pollution on wildlife is scarce and refers to very few species. It is very difficult to estimate from the available data the total doses that can accumulate during specific stages of the life cycle (embryonic development or reproductive age), in changes in behavior or depending on environmental factors such as seasonality.[17] The phenomena of radioactive bioaccumulation, bioconcentration and biomagnification, however, are especially known to sea level. They are caused by the recruitment and retention of radioisotopes by bivalves, crustaceans, corals and phytoplankton, which then amounted to the rest of the food chain at low concentration factors.[18]
Radiobiological literature and IAEA establish a safe limit of absorbed dose of 0.001 Gy/d for terrestrial animals and 0.01 Gy/d for plants and marine biota, although this limit should be reconsidered for long-lived species with low reproductive capacity.[19]

Radiation tests in model organisms that determine the effects of high radiation on animals and plants are:[19]
- Chromosomal aberrations.
- DNA damage.
- Cancer, particularly leukemia.[3]
- Leukopenia.
- Growth reduction.
- Reproductive deficiencies: sterility, reduction in fecundity, and occurrence of developmental abnormalities or reduction in viability of offspring
- Reduced seed germination.
- Burned tissues exposed to radiation.
- Mortality, including both acute lethality and long-term reduction in life span.
The effects of radioactivity on bacteria are given, as in eukaryotes, by ionization of water and production of reactive oxygen species. These compounds mutate DNA strands and produce genetic damage, inducing newly lysis and subsequent cell death.[21][22]
Its action on viruses, on the other hand, results in damaged nucleic acids and viral inactivation.[23] They have a sensory threshold ranging between 1000 and 10,000 Gy (range occupying most biological organisms) which decreases with increasing genome size.[24]
Bacterial bioremediation
The biochemical transformation of radionuclides into stable isotopes by bacterial species varies metabolism of organic compounds from carbon sources. They are highly energetic radioactive forms which can be converted indirectly by the process of microbial energy transfer.[2]
Radioisotopes can be transformed directly through changes in valence state by acting as acceptors or by acting as cofactors to enzymes. They can also be transformed indirectly by reducing and oxidizing agents produced by microorganisms that cause changes in pH or redox potential. Other processes include precipitation and complexation of surfactants, or chelating agents that bind to radioactive elements. Human intervention, on the other hand, can improve these processes through genetic engineering and omics, or by injection of microorganisms or nutrients into the treatment area.[2][6]
Bioreduction
According to the radioactive element and the specific site conditions, bacteria can enzymatically immobilize radionuclides directly or indirectly. Their redox potential is exploited by some microbial species to carry out reductions that alter the solubility and hence, mobility, bioavailability and radiotoxicity. This waste treatment technique called bioreduction or enzymatic biotransformation is very attractive because it can be done in mild conditions for the environment, does not produce hazardous secondary waste and has potential as a solution for waste of various kinds.[5]
Direct enzymatic reduction is the change of radionuclides of a higher oxidation state to a lower one made by facultative and obligate anaerobes. The radioisotope interact with binding sites of metabolically active cells and is used as terminal electron acceptor in the electron transport chain where compounds such as ethyl lactate act as electron donors under anaerobic respiration.[5]
The periplasm plays a very important role in these bioreductions. In the reduction of uranium (VI) to insoluble uranium (IV), made by Shewanella putrefaciens, Desulfovibrio vulgaris, Desulfovibrio desulfuricans and Geobacter sulfurreducens, the activity of periplasmic cytochromes is required. The reduction of technetium (VII) to technetium (IV) made by S. putrefaciens, G. sulfurreducens, D. desulfuricans, Geobacter metallireducens and Escherichia coli, on the other hand, requires the presence of the complex formate hydrogenlyase, also placed in this cell compartment.[3]
Other radioactive actinides such as thorium, plutonium, neptunium and americium are enzymatically reduced by Rhodoferax ferrireducens, S. putrefaciens and several species of Geobacter, and directly form an insoluble mineral phase.[3]
The phenomenon of indirect enzymatic reduction is carried out by sulfate-reducing and dissimilatory metal-reducing bacteria on excretion reactions of metabolites and breakdown products. There is a coupling of the oxidation of organic acids —produced by the excretion of these heterotrophic bacteria— with the reduction of iron or other metals and radionuclides, which forms insoluble compounds that can precipitate as oxide and hydroxide minerals. In the case of sulfate-reducing bacteria hydrogen sulfide is produced, promoting increased solubility of polluting radionuclides and their bioleaching (as liquid waste that can then be recovered).[3][5]
There are several species of reducing microorganisms that produce indirect sequestering agents and specific chelators, such as siderophores. These sequestering agents are crucial in the complexation of radionuclides and increasing their solubility and bioavailability. Microbacterium flavescens, for example, grows in the presence of radioisotopes such as plutonium, thorium, uranium or americium and produces organic acids and siderophores that allow the dissolution and mobilization of radionuclides through the soil. It seems that siderophores on bacterial surface could also facilitate the entry of these elements within the cell as well. Pseudomonas aeruginosa also secretes chelating agents out that meet uranium and thorium when grown in a medium with these elements. In general, it has also been found that enterobactin siderophores are extremely effective in solubilizing actinide oxides of plutonium.[3][5]
Citrate complexes
Citrate is a chelator which binds to certain transition metals and radioactive actinides. Stable complexes such as bidentate, tridentate (ligands with more than one atom bound) and polynuclear complexes (with several radioactive atoms) can be formed with citrate and radionuclides, which receive a microbial action. Anaerobically, Desulfovibrio desulfuricans and species of Shewanella and Clostridium genus are able to reduce bidentate complexes of uranyl-citrate (VI) to uranyl-citrate (IV) and make them precipitate, despite not being able to degrade metabolically complexed citrate at the end of the process.[3] In denitrifying and aerobic conditions, however, it has been determined that it is not possible to reduce or degrade these uranium complexes. Bioreduction do not get a head when they are citrate complex mixed metal complexes or when they are tridentate, monomeric or polynuclear complexes, since they become recalcitrant and persistent in the environment.[5][26] From this knowledge exists a system that combines the degradation of radionuclide-citrate complex with subsequent photodegradation of remaining reduced uranyl-citrate (previously not biodegradated but sensitive to light), which allows for stable precipitates of uranium and also of thorium, strontium or cobalt from contaminated lands.[5]
Biosorption, bioaccumulation and biomineralization
.png)
The set of strategies that comprise biosorption, bioaccumulation and biomineralization are closely related to each other, because one way or another have a direct contact between the cell and radionuclide. These mechanisms are evaluated accurately using advanced analysis technologies such as electron microscopy, X-ray diffraction and XANES, EXAFS and X-ray spectroscopies.[2][27]
Biosorption and bioaccumulation are two metabolic actions that are based on the ability to concentrate radionuclides over a thousand times the concentration of the environment. They consist of complexation of radioactive waste with phosphates, organic compounds and sulfites so that they become insoluble and less exposed to radiotoxicity. They are particularly useful in biosolids for agricultural purposes and soil amendments, although most properties of these biosolids are unknown.[28]
Biosorption method is based on passive sequestration of positively charged radioisotopes by lipopolysaccharides (LPS) on the cell membrane (negatively charged), either live or dead bacteria. Its efficiency is directly related to the increase in temperature and can last for hours, being a much faster method than direct bioreduction. It occurs through the formation of slimes and capsules, and with a preference for binding to the phosphate and phosphoryl groups (although it also occurs with carboxyl, amine or sulfhydryl groups). Firmicutes and other bacteria like Citrobacter freudii have significant biosorption capabilities; Citrobacter does it through electrostatic interaction of uranium with phosphates of their LPS.[3][4]
Quantitative analyzes determine that, in the case of uranium, biosorption may vary within a range between 45 and 615 milligrams per gram of cell dry weight. However, it is a technique that requires a high amount of biomass to affect bioremediation; it presents problems of saturation and other cations that compete for binding to the bacterial surface.[4]
Bioaccumulation refers to uptake of radionuclides into the cell, where they are retained by complexations with negatively charged intracellular components, precipitation or granules formations. Unlike biosorption, this is an active process: it depends on an energy-dependent transport system.[25] Some metals or radionuclides can be absorbed by bacteria accidentally because of its resemblance to dietary elements for metabolic pathways. Several radioisotopes of strontium, for example, are recognized as analogs of calcium and incorporated within Micrococcus luteus.[5] Uranium, however, has no known function and is believed that its entry into the cell interior may be due to its toxicity (it is able to increase membrane permeability).[4]

Furthermore, biomineralization —also known as bioprecipitation— is the precipitation of radionuclides through the generation of microbial ligands, resulting in the formation of stable biogenic minerals. These minerals have a very important role in the retention of radioactive contaminants. A very localized and produced enzymatically ligand concentration is involved and provides a nucleation site for the onset of biomineral precipitation.[29] This is particularly relevant in precipitations of phosphatase activity-derivate biominerals, which cleavage molecules such as glycerol phosphate on periplasm. In Citrobacter and Serratia genera, this cleavage liberates inorganic phosphates (HPO42−) that precipitates with uranyl ion (UO22+) and cause deposition of polycrystalline minerals around the cell wall.[3][30] Serratia also form biofilms that promote precipitation of chernikovite (rich in uranium) and additionally, remove up to 85% of cobalt-60 and 97% of cesium-137 by proton substitution of this mineral.[27] In general, biomineralization is a process in which the cells do not have limitations of saturation and can accumulate up to several times its own weight as precipitated radionuclides.[5]
Investigations of terrestrial and marine bacterial isolates belonging to the genera Aeromonas, Bacillus, Myxococcus, Pantoea, Pseudomonas, Rahnella and Vibrio have also demonstrated the removal of uranium radioisotopes as phosphate biominerals in both oxic and anoxic growth conditions.[27]
Biostimulation and bioaugmentation
Aside from bioreduction, biosorption, bioaccumulation and biomineralization, which are bacterial strategies for natural attenuation of radioactive contamination, there are also human methods that increase the efficiency or speed of microbial processes. This accelerated natural attenuation involves an intervention in the contaminated area to improve conversion rates of radioactive waste, which tend to be slow. There are two variants: biostimulation and bioaugmentation.[32]
Biostimulation is the addition of nutrients with trace elements, electron donors or electron acceptors to stimulate activity and growth of natural indigenous microbial communities.[5][32] It can range from simple fertilization or infiltration (called passive biostimulation) to more aggressive injections to the ground, and is widely used at US DOE sites.[28] Nitrate is used as nutrient to biostimulate the reduction of uranium, because it serves as very energetically favorable electron acceptor for metal-reducing bacteria. However, many of these microorganisms (Geobacter, Shewanella or Desulfovibrio) exhibit resistance genes to heavy metals that limit their ability to bioremediate radionuclides. In these particular cases, a carbon source such as ethanol is added to the medium to promote the reduction of nitrate at first, and then of uranium. Ethanol is also used in soil injection systems with hydraulic recirculations: it raises the pH and promotes the growth of denitrifying and radionuclide-reducing bacteria, that produce biofilms and achieve almost 90% decrease in the concentration of radioactive uranium.[3]
A number of geophysical techniques have been used to monitor the effects of in situ biostimulation trials including measurement of: spectral ionization potential, self potentials, current density, complex resistivity and also reactive transport modelling (RTM), which measures hydrogeological and geochemical parameters to estimate chemical reactions of the microbial community.[4]
Bioaugmentaton, on the other hand, is the deliberated addition to the environment of microorganisms with desired traits to accelerate bacterial metabolic conversion of radioactive waste. They are often added when necessary species for bioremediation do not exist in the treatment place.[5][32] This technique has shown in field trials over the years that it does not offer better results than biostimulation; neither it is clear that introduced species can be distributed effectively through the complex geological structures of most subsurface environments or that can compete long term with the indigenous microbiota.[2][28]
Genetic engineering and omics
Omics, especially genomics and proteomics, allow identifying and evaluating genes, proteins and enzymes involved in radionuclide bioremediation, apart from the structural and functional interactions that exist between them and other metabolites. Genome sequencing of various microorganisms has uncovered, for example, that Geobacter sulfurreducens possess more than 100 coding regions for c-type cytochromes involved in bioremediation radionuclide, or that NiCoT gene is significantly overexpressed in Rhodopseudomonas palustris and Novosphingobium aromaticivorans when grown in medium with radioactive cobalt.[2][3]
From this information, different genetic engineering and recombinant DNA techniques are being developed to generate specific bacteria for bioremediation. Some constructs expressed in microbial species are phytochelatins, polyhistidines and other polypeptides by fusion-binding domains to outer-membrane-anchored proteins.[3] Some of these genetically modified strains are derived from Deinococcus radiodurans, one of the most radiation-resistant organisms. D. radiodurans is capable to resist oxidative stress and DNA damage from radiation, and reduces technetium, uranium and chromium naturally as well. Besides, through insertion of genes from other species it has been achieved that it can also precipitates uranyl phosphates and degrades mercury by using toluene as an energy source to grow and stabilize other priority radionuclides.[2][4]
Directed evolution of bacterial proteins related to bioremediation of radionuclides is also a field research. YieF enzyme, for example, naturally catalyzes the reduction of chromium with a very wide range of substrates. Following protein engineering, however, it has also been able to participate in uranyl ion reduction.[33]
Plant bioremediation
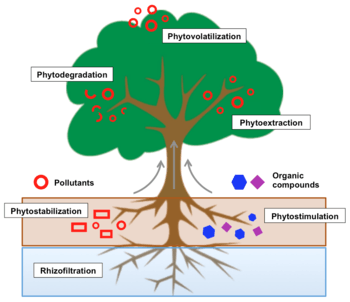
The use of plants to remove contaminants from the environment or to render them less harmful is called phytoremediation. In the case of radionuclides, it is a viable technology when decontamination times are long and waste are scattered at low concentrations.[34][35]
Some plant species are able to transform the state of radioisotopes (without suffering toxicity) concentrating them in different parts of their structure, making them rush through the roots, making them volatile or stabilizing them on the ground. As in bacteria, plant genetic engineering procedures and biostimulation —called phytostimulation— have improved and accelerate these processes, particularly with regard to fast-growing plants.[35] The use of Agrobacterium rhizogenes, for example, is quite widespread and significantly increases radionuclide uptake by the roots.[36]
Phytoextraction
In phytoextraction (also phytoaccumulation, phytosequesteration or phytoabsorption)[37] plants carry radioactive waste from the root system to the vascular tissue and become concentrated in the biomass of shoots. It is a technique that removes radionuclides without destroying the soil structure, with minimal impact on soil fertility and valid for large areas with a low level of radioactivity. Its efficiency is evaluated through bioaccumulation coefficient (BC) or total removal of radionuclides per m2, and is proven to attract cesium-137, strontium-90, technetium-99, cerium-144, plutonium-240, americium-241, neptunium-237 and various radioisotopes of thorium and radium.[35] By contrast, it requires large biomass production in short periods of time.[36]
Species like common heather or amaranths are able to concentrate cesium-137, the most abundant radionuclide in the Chernobyl Exclusion Zone. In this region of Ukraine, mustard greens could remove up to 22% of average levels of cesium activity in a single growing season. In the same way, bok choy and mustard greens can concentrate 100 times more uranium than other species.[35]
Rhizofiltration

Rhizofiltration is the adsorption and precipitation of radionuclides in plant roots or absorption thereof if soluble in effluents. It has great efficiency in the treatment of cesium-137 and strontium-90, particularly by algae and aquatic plants, such as Cladophora and Elodea genera, respectively. It is the most efficient strategy for bioremediation technologies in wetlands,[37] but must have a continuous and rigorous control of pH to make it an optimal process.[36]
From this process, some strategies have been designed based on sequences of ponds with a slow flow of water to clean polluted water with radionuclides. The results of these facilities, for flows of 1000 liters of effluent are about 95% retention of radiation in the first pond (by plants and sludge), and over 99% in three-base systems.[35]
The most promising plants for rhizofiltration are sunflowers. They are able to remove up to 95% of uranium of contaminated water in 24 hours, and experiments in Chernobyl have demonstrated that they can concentrate on 55 kg of plant dry weight all the cesium and strontium radioactivity from an area of 75 m2 (stabilized material suitable for transfer to a nuclear waste repository).[35]
Phytovolatilization
Phytovolatilization involves the capture and subsequent transpiration of radionuclides into the atmosphere. It does not remove contaminants but releases them in volatile form (less harmful). Despite not having too many applications for radioactive waste, it is very useful for the treatment of tritium, because it exploits plants' ability to transpire enormous amounts of water.[35][37]
The treatment applied to tritium (shielded by air produces almost no external radiation exposure, but its incorporation in water presents a health hazard when absorbed into the body) uses polluted effluents to irrigate phreatophytes. It becomes a system with a low operation cost and low maintenance, with savings of about 30% in comparison to conventional methods of pumping and covering with asphalt.[35]
Phytostabilization
Phytostabilization is an specially valid strategy for radioactive contamination based on the immobilization of radionuclides in the soil by the action of the roots. This can occur by adsorption, absorption and precipitation within root zone, and ensures that radioactive waste can not be dispersed because soil erosion or leaching. It is useful in controlling tailings from strip and open pit uranium mines, and guarantees to retrieve the ecosystem.[35][37] However, it has significant drawbacks such as large doses of fertilizer needed to reforest the area, apart from radioactive source (which implies long-term maintenance) remaining at the same place.[36]
Fungal bioremediation

Several fungi species have radioactive resistance values equal to or greater than more radioresistant bacteria; they perform mycoremediation processes. It was reported that some fungi had the ability of growing into, feeding, generating spores and decomposing pieces of graphite from destroyed reactor No. 4 at the Chernobyl Nuclear Power Station, which is contaminated with high concentrations of cesium, plutonium and cobalt radionuclides. They were called radiotrophic fungi.[38]
Since then, it has been shown that some species of Penicillium, Cladosporium, Paecilomyces and Xerocomus are able to use ionizing radiation as energy through the electronic properties of melanins.[38][39] In their feeding they bioaccumulate radioisotopes, creating problems on concrete walls of deep geological repositories.[40] Other fungi like oyster mushrooms can bioremediate plutonium-239 and americium-241.[41]
Ways of research
Current research on bioremediation techniques is fairly advanced and molecular mechanisms that govern them are well known. There are, however, many doubts about the effectiveness and possible adversities of these processes in combination with the addition of agrochemicals. In soils, the role of mycorrhizae on radioactive waste is poorly described and sequestration patterns of radionuclides are not known with certainty.[42]
Longevity effects of some bacterial processes, such as maintenance of uranium in insoluble form because of bioreductions or biomineralizations, are unknown. There are not clear details about the electronic transfer from some radionuclides with these bacterial species either.[4]
Another important aspect is the change of ex situ or laboratory scale processes to their real application in situ, in which soil heterogeneity and environmental conditions generate reproduction deficiencies of optimal biochemical status of the used species, a fact that decreases the efficiency. This implies finding what are the best conditions in which to carry out an efficient bioremediation with anions, metals, organic compounds or other chelating radionuclides that can compete with the uptake of interest radioactive waste.[3] Nevertheless, in many cases research is focused on the extraction of soil and water and its ex situ biological treatment to avoid these problems.[5]
Finally, the potential of GMOs is limited by regulatory agencies in terms of responsibility and bioethical issues. Their release require support on the action zone and comparability with indigenous species. Multidisciplinary research is focused on defining more precisely necessary genes and proteins to establish new free-cell systems, which may avoid possible side effects on the environment by the intrusion of transgenic or invasive species.[3]
See also
References
- ↑ Idaho National Laboratory. "The INL & The Snake River Plain Aquifer" (PDF). Office of Environmental Management. Washington: United States Department of Energy.
- 1 2 3 4 5 6 7 Faison, B; McCullough, J; Hazen, TC; Benson, SM; Palmisano, A (2003). A NABIR Primer, ed. Bioremediation of metals and radionuclides: What it is and how it works (PDF) (2nd edition. Revision by the Lawrence Berkeley National Laboratory ed.). Washington: United States Department of Energy.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 Prakash, D; Gabani, P; Chandel, A.K; Ronen, Z; Singh, O.V (2013). "Bioremediation: a genuine technology to remediate radionuclides from the environment" (PDF). Microbial Biotechnology. New York: John Wiley & Sons. 6 (4): 349–360. doi:10.1111/1751-7915.12059. PMC 3917470
 .
. - 1 2 3 4 5 6 7 8 Newsome, L; Morris, K; Lloyd, J.R (2014). "The biogeochemistry and bioremediation of uranium and other priority radionuclides" (PDF). Chemical Geology. Elsevier. 363: 164–184. doi:10.1016/j.chemgeo.2013.10.034.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 Francis, A.J; Nancharaiah, Y.V (2015). "9. In situ and ex situ bioremediation of radionuclide-contaminated soils at nuclear and NORM sites". In van Velzen, L. Environmental Remediation and Restoration of Contaminated Nuclear and Norm Sites (PDF). Woodhead Publishing Series in Energy. Elsevier. pp. 185–236. doi:10.1016/B978-1-78242-231-0.00009-0. ISBN 978-1-78242-231-0.
- 1 2 Francis, A.J (2006). Microbial Transformations of Radionuclides and Environmental Restoration Through Bioremediation (PDF). Symposium on "Emerging Trends in Separation Science and Technology". Mumbai: Brookhaven National Laboratory.
- ↑ Consejo de Seguridad Nuclear. Ministerio de Industria, Turismo y Comercio de España, ed. "Radiación natural y artificial" (Web) (in Spanish). Retrieved 24 February 2016.
- ↑ Varskog, P; Stralberg, E; Varskog, A.T.S; Raaum, A (2003). Naturally occurring radionuclides in the marine environment: an overview of current knowledge with emphasis on the North Sea area (PDF). Kjeller: Norse Decom AS. p. 7. ISBN 82-92538-01-1.
- 1 2 3 Idaho State University (ed.). "Radioactivity in Nature" (Web). Retrieved 25 February 2016.
- 1 2 Hu, Q; Weng, J; Wang, J (2010). "Sources of anthropogenic radionuclides in the environment: a review" (Web). Journal of Environmental Radioactivity. Arlington: Elsevier. 101 (6): 426–437. doi:10.1016/j.jenvrad.2008.08.004.
- ↑ Campbell, J.A (1983). "44.4.1. Tritium". In Riley, J.P; Chester, R. Chemical Oceanography (Web). 8. New York: Academic Press. pp. 111–117. ISBN 9781483219837.
- ↑ Aoyama, M; Hirose, K (2008). "Radiometric determination of anthropogenic radionuclides in seawater" (Web). Radioactivity in the Environment. Elsevier. 11: 137–162. doi:10.1016/S1569-4860(07)11004-4. ISSN 1569-4860.
- ↑ Riley, R.G; Zachara, J.M; Wobber, F.J (1992). "Chemical Contaminants on DOE Lands and Selection of Contaminant Mixtures for Subsurface Science Research" (PDF). Office of Energy Research. United States Department of Energy: 22.
- ↑ Vienna International Centre (2009). "Classification of radioactive waste: general safety guide" (PDF). IAEA safety standards series. Viena: International Atomic Energy Agency (nº GSG-1): 5–6. ISBN 9789201092090. ISSN 1020-525X.
- 1 2 Sharma, B.K; Sharma, A; Sharma, M (2007). "Effects of radioactive pollution". In Sharma, B.K. Environmental Chemistry (Web). Meerut: Krishna Prakashan Media. ISBN 9788182830127.
- ↑ Brenner, D.J; Doll, R; Goodhead, D.T; Hall, E.J; Land, C.E (2003). "Cancer risks attributable to low doses of ionizing radiation: Assessing what we really know" (PDF). PNAS. National Academy of Sciences. 100 (24): 13761–13766. doi:10.1073/pnas.2235592100. ISSN 1091-6490.
- ↑ Linsley, G (1997). "Radiation & the environment: Assessing effects on animals and plants" (PDF). IAEA Bulletin. International Atomic Energy Agency.
- ↑ Stewart, G.M; Fowler, S.W; Fisher, N.S (2011). "8. The bioaccumulation of U- and Th- series radionuclides in marine organisms". In Krishnaswami, S; Cochran, J.K. U-Th Series Nuclides in Aquatic Systems (Web). Radioactivity in the Environment. 13. Amsterdam: Elsevier. pp. 269–305. ISBN 9780080564883. ISSN 1569-4860.
- 1 2 Barnthouse, L.W (1995). Environmental Sciences Division, ed. "Effects of ionizing radiation on terrestrial plants and animals: a workshop report" (PDF) (4496). Tennessee: United States Department of Energy.
- ↑ Gager, C.S (1909). "The Influence of Radium Rays on a Few Life Processes of Plants" (Djvu). The Popular Science Monthly. New York: The Science Press. 74: 222–232.
- ↑ Confalonieri, F; Sommer, S (2011). "Bacterial and archaeal resistance to ionizing radiation" (PDF). Journal of Physics: Conference Series. Orsay: IOP Publishing. 261. doi:10.1088/1742-6596/261/1/012005.
- ↑ Kelner, A; Dexter Bellamy, W; Stapleton, G.E; Zelle, M.R (1955). "Symposium on radiation effects on cells and bacteria" (PDF). Bacteriological Reviews. American Society of Biology. 19 (1): 22–24. PMC 180808
 .
. - ↑ Specter, S; Jeffries, D (1996). "18. Disinfection". In W.J Mahy, B; O Krango, H. Virology Methods Manual (Web). San Diego: Academic Press. pp. 353–356. ISBN 9780080543581.
- ↑ Lowy, R.J (2005). "Ionizing Radiation Inactivation of Medically Relevant Viruses". In Gazsó, L.G; Ponta, C.C. Radiation Inactivation of Bioterrorism Agents (Web). NATO Science Series. Series I, Life and behavioural sciences. 365. Budapest: IOS Press. pp. 175–186. ISBN 9781586034887. ISSN 1566-7693.
- 1 2 Mtimunye, P.J; Chirwa, E.M.N (2013). "Chapter 8: Bioremediation of Radiotoxic Elements under Natural Environmental Conditions". In Yogesh, B.P; Rao, P. Applied Bioremediation - Active and Passive Approaches (Web). Intech. doi:10.5772/56909. ISBN 978-953-51-1200-6.
- ↑ Francis, A.J (2012). "6. Impacts of microorganisms on radionuclides in contaminated environments and waste materials". In Poinssot, C; Geckeis, H. Radionuclide Behaviour in the Natural Environment: Science, Implications and Lessons for the Nuclear industry. Woodhead Publishing Series in Energy. Woodhead Publishing. pp. 161–226. ISBN 9780857097194.
- 1 2 3 Martinez, R.J; Beazley, M.J; Sobecky, P.A (2014). "Phosphate-Mediated Remediation of Metals and Radionuclides" (Web). Advances in Ecology. Hindawi Publishing. doi:10.1155/2014/786929.
- 1 2 3 Hazen, T.C; Tabak H.H (2005). "Developments in bioremediation of soils and sediments polluted with metals and radionuclides: 2. Field research on bioremediation of metals and radionuclides" (PDF). Reviews in Environmental Science and Bio/Technology. Springer. 4 (3): 157–183. doi:10.1007/s11157-005-2170-y. ISSN 1572-9826.
- ↑ Walther, C; Guptha, D.K (2015). "2.3 Biomineralization/Bioprecipitation". Radionuclides in the Environment: Influence of chemical speciation and plant uptake on radionuclide migration (Web). Switzerland: Springer. p. 178. ISBN 9783319221717.
- ↑ Mackasie, L.E; Young, P; Patterson-Beedle, M (2004). "24. Bacterial precipitation of metal phosphates". In Valsami-Jones, E. Phosphorus in Environmental Technologies: Principles and Applications (Web). Integrated environmental technology series. London: International Water Publishing. pp. 553–557. ISBN 9781843390015.
- ↑ Chang, Y (2005). "In Situ Biostimulation of Uranium Reducing Microorganisms at the Old Rifle UMTRA Site" (Web). Knoxville: University of Tennessee.
- 1 2 3 Natural and Accelerated Bioremediation Research. United States Department of Energy, ed. "II. Program Goals and Management Strategy" (Web). Retrieved 14 May 2015.
- ↑ Matin, A.C (2006). Stanford University, ed. "Development of combinatorial bacteria for metal and radionuclide bioremediation" (PDF). Stanford: DOE's Office of Scientific and Technical Information.
- ↑ Willey, N; Collins, C (2007). "Phytoremediation of soils contaminated with radionuclides". In Shaw, G. Radioactivity in the Terrestrial Environment (Web). 10. Elsevier. pp. 43–69. doi:10.1016/S1569-4860(06)10003-0. ISBN 9780080474892.
- 1 2 3 4 5 6 7 8 9 Dushenkov, S (2003). "Trends in phytoremediation of radionuclides" (Web). Plant and Soil. Netherlands: Kluwer Academic Publishers. 249 (1): 167–175. doi:10.1023/A:1022527207359. ISSN 1573-5036.
- 1 2 3 4 Celso de Mello-Farias, P; Soares, A.L; Leoneti, C (2011). "Chapter 16. Transgenic Plants for Enhanced Phytoremediation – Physiological Studies". In Alvarez, M. Genetic Transformation (Web). Intech. doi:10.5772/24355. ISBN 978-953-307-364-4.
- 1 2 3 4 Kumar, D; Walther, C, eds. (2014). "4. Phytoremediation techniques". Radionuclide Contamination and Remediation Through Plants. Hannover: Springer. pp. 9–14. ISBN 9783319076652.
- 1 2 Dadachova, E; Casadevall, A (2008). "Ionizing Radiation: how fungi cope, adapt, and exploit with the help of melanin" (PDF). Current Opinion in Microbiology. Elsevier. 11 (6): 525–531. doi:10.1016/j.mib.2008.09.013. PMC 2677413
 .
. - ↑ Kalac, P (2001). "A review of edible mushroom radioactivity" (Web). Food Chemistry. Elsevier. 75 (1): 29–35. doi:10.1016/S0308-8146(01)00171-6.
- ↑ Fomina, M; Burford, E.P; M. Gadd, G (2006). "Fungal dissolution and transformation of minerals: significance for nutrient and metal mobility". In Gadd, G.M. Fungi in Biogeochemical Cycles (Web). Cambridge University Press. pp. 236–266. doi:10.1017/CBO9780511550522.011. ISBN 9780521845793.
- ↑ Galanda, D; Mátel, L; Strišovská, J; Dulanská, S (2014). "Mycoremediation: the study of transfer factor for plutonium and americium uptake from the ground" (Web). Journal of Radioanalytical and Nuclear Chemistry. Budapest: Springer. 299 (3): 1411–1416. doi:10.1007/s10967-013-2909-9. ISSN 1588-2780.
- ↑ Zhu, Y.G; Shaw, G (2000). "Soil contamination with radionuclides and potential remediation". Chemosphere. Elsevier. 41 (1-2): 121–128. doi:10.1016/S0045-6535(99)00398-7. PMID 10819188.
External links
| Wikimedia Commons has media related to Environmental restoration. |
- Bioremediation of radioactive waste. (PDF) Scientific poster of the Bachelor Thesis related to this article. Autonomous University of Barcelona Digital Repository of Documents.
| Library resources about Bioremediation of radioactive waste |
.png)
