Human microbiota
The human microbiota is the aggregate of microorganisms, a microbiome that resides on or within a number of tissues and biofluids, including the skin, mammary glands, placenta, seminal fluid, uterus, ovarian follicles, lung, saliva, oral mucosa, conjunctiva, and gastrointestinal tracts. They include bacteria, fungi, and archaea. Micro-animals which live on the human body are excluded. The human microbiome refers to their genomes.[1]
Humans are colonized by many microorganisms; the traditional estimate was that humans live with ten times more non-human cells than human cells; more recent estimates have lowered that to 3:1 and even to approximately the same number; all the numbers are estimates.[2][3][4][5] Some microbiota that colonize humans have not merely a commensal (a non-harmful coexistence), but rather a mutualistic relationship with their human hosts.[1]:700[6] Conversely, some non-pathogenic microbiota can harm human hosts via the metabolites that they produce, like trimethylamine.[7][8] Certain microbiota perform tasks that are known to be useful for the human host; for most, the role is not well understood. Those that are expected to be present, and that under normal circumstances do not cause disease, are deemed normal flora or normal microbiota.[1]
The Human Microbiome Project took on the project of sequencing the genome of the human microbiota, focusing particularly on the microbiota that normally inhabit the skin, mouth, nose, digestive tract, and vagina.[1] It reached a milestone in 2012 when it published initial results.[9]
Terminology
Though widely known as flora or microflora, this is a misnomer in technical terms, since the word root flora pertains to plants, and biota refers to the total collection of organisms in a particular ecosystem. Recently, the more appropriate term microbiota is applied, though its use has not eclipsed the entrenched use and recognition of flora with regard to bacteria and other microorganisms. Both terms are being used in different literature.[6]
Relative numbers
As of 2014, it was often reported in popular media and in the scientific literature that there are about 10 times as many microbial cells in the human body than there are human cells; this figure was based on estimates that the human microbiome includes around 100 trillion bacterial cells and an adult human typically has around 10 trillion human cells.[2] In 2014 the American Academy of Microbiology published an FAQ that emphasized that the number of microbial cells and the number of human cells are both estimates, and noted that recent research had arrived at a new estimate of the number of human cells at around 37 trillion cells, meaning that the ratio of microbial to human cells is probably about 3:1.[2][3] In 2016 another group published a new estimate of ratio as being roughly 1:1 (1.3:1, with "an uncertainty of 25% and a variation of 53% over the population of standard 70 kg males").[4][5]
Study
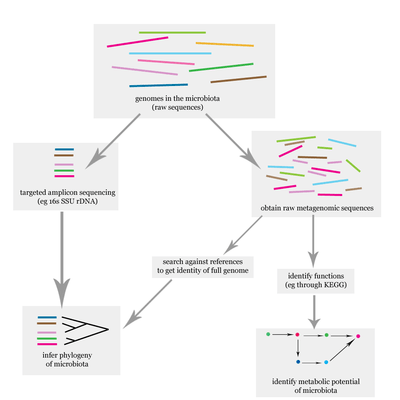
The problem of elucidating the human microbiome is essentially identifying the members of a microbial community which includes bacteria, eukaryotes, and viruses.[10] This is done primarily using DNA-based studies, though RNA, protein and metabolite based studies are also performed.[10][11] DNA-based microbiome studies typically can be categorized as either targeted amplicon studies or more recently shotgun metagenomic studies. The former focuses on specific known marker genes and is primarily informative taxonomically, while the latter is an entire metagenomic approach which can also be used to study the functional potential of the community.[10] One of the challenges that is present in human microbiome studies but not in other metagenomic studies is to avoid including the host DNA in the study.[12]
Aside from simply elucidating the composition of the human microbiome, one of the major questions involving the human microbiome is whether there is a "core", that is, whether there is a subset of the community that is shared among most humans.[13][14] If there is a core, then it would be possible to associate certain community compositions with disease states, which is one of the goals of the Human Microbiome Project. It is known that the human microbiome is highly variable both within a single subject and between different individuals. For example, the gut microbiota of humans is markedly dissimilar between individuals, a phenomenon which is also observed in mice.[6]
On 13 June 2012, a major milestone of the Human Microbiome Project (HMP) was announced by the NIH director Francis Collins.[9] The announcement was accompanied with a series of coordinated articles published in Nature[15][16] and several journals in the Public Library of Science (PLoS) on the same day. By mapping the normal microbial make-up of healthy humans using genome sequencing techniques, the researchers of the HMP have created a reference database and the boundaries of normal microbial variation in humans. From 242 healthy U.S. volunteers, more than 5,000 samples were collected from tissues from 15 (men) to 18 (women) body sites such as mouth, nose, skin, lower intestine (stool), and vagina. All the DNA, human and microbial, were analyzed with DNA sequencing machines. The microbial genome data were extracted by identifying the bacterial specific ribosomal RNA, 16S rRNA. The researchers calculated that more than 10,000 microbial species occupy the human ecosystem and they have identified 81 – 99% of the genera.
Types
Bacteria
Populations of microbes (such as bacteria and yeasts) inhabit the skin and mucosal surfaces in various parts of the body. Their role forms part of normal, healthy human physiology, however if microbe numbers grow beyond their typical ranges (often due to a compromised immune system) or if microbes populate (such as through poor hygiene or injury) areas of the body normally not colonized or sterile (such as the blood, or the lower respiratory tract, or the abdominal cavity), disease can result (causing, respectively, bacteremia/sepsis, pneumonia, and peritonitis).
The Human Microbiome Project found that individuals host thousands of bacterial types, different body sites having their own distinctive communities. Skin and vaginal sites showed smaller diversity than the mouth and gut, these showing the greatest richness. The bacterial makeup for a given site on a body varies from person to person, not only in type, but also in abundance. Bacteria of the same species found throughout the mouth are of multiple subtypes, preferring to inhabit distinctly different locations in the mouth. Even the enterotypes in the human gut, previously thought to be well-understood, are from a broad spectrum of communities with blurred taxon boundaries.[17][18]
It is estimated that 500 to 1,000 species of bacteria live in the human gut but belong to just a few phyla: Firmicutes and Bacteroidetes dominate but there are also Proteobacteria, Verrumicrobia, Actinobacteria, Fusobacteria and Cyanobacteria.[19]
A number of types of bacteria, such as Actinomyces viscosus and A. naeslundii, live in the mouth, where they are part of a sticky substance called plaque. If this is not removed by brushing, it hardens into calculus (also called tartar). The same bacteria also secrete acids that dissolve tooth enamel, causing tooth decay.
The vaginal microflora consist mostly of various lactobacillus species. It was long thought that the most common of these species was Lactobacillus acidophilus, but it has later been shown that L. iners is in fact most common, followed by L. crispatus. Other lactobacilli found in the vagina are L. jensenii, L. delbruekii and L. gasseri. Disturbance of the vaginal flora can lead to infections such as bacterial vaginosis or candidiasis ("yeast infection").
Archaea
Archaea are present in the human gut, but, in contrast to the enormous variety of bacteria in this organ, the numbers of archaeal species are much more limited.[20] The dominant group are the methanogens, particularly Methanobrevibacter smithii and Methanosphaera stadtmanae.[21] However, colonization by methanogens is variable, and only about 50% of humans have easily detectable populations of these organisms.[22]
As of 2007, no clear examples of archaeal pathogens were known,[23][24] although a relationship has been proposed between the presence of some methanogens and human periodontal disease.[25]
Fungi
Fungi, in particular yeasts, are present in the human gut.[26][27][28][29] The best-studied of these are Candida species due to their ability to become pathogenic in immunocompromised and even in healthy hosts.[27][28][29] Yeasts are also present on the skin,[26] such as Malassezia species, where they consume oils secreted from the sebaceous glands.[30][31]
Viruses
Viruses, especially bacterial viruses (bacteriophages), colonize various body sites. These colonized sites include the skin,[32] gut,[33] lungs,[34] and oral cavity.[35] Virus communities have been associated with some diseases, and do not simply reflect the bacterial communities.[36][37][38]
Anatomical areas
Skin
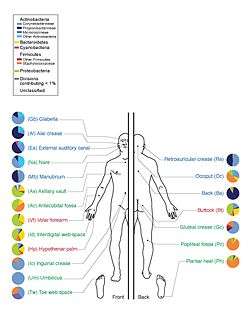
A study of twenty skin sites on each of ten healthy humans found 205 identified genera in nineteen bacterial phyla, with most sequences assigned to four phyla: Actinobacteria (51.8%), Firmicutes (24.4%), Proteobacteria (16.5%), and Bacteroidetes (6.3%).[39] A large number of fungal genera are present on healthy human skin, with some variability by region of the body; however, during pathological conditions, certain genera tend to dominate in the affected region.[26] For example, Malassezia is dominant in atopic dermatitis and Acremonium is dominant on dandruff-afflicted scalps.[26]
The skin acts as a barrier to deter the invasion of pathogenic microbes. The human skin contains microbes that reside either in or on the skin and can be residential or transient. Resident microorganism types vary in relation to skin type on the human body. A majority of microbes reside on superficial cells on the skin or prefer to associate with glands. These glands such as oil or sweat glands provide the microbes with water, amino acids, and fatty acids. In addition, resident bacteria that associated with oil glands are often Gram-positive and can be pathogenic.[1]
Conjunctiva
A small number of bacteria and fungi are normally present in the conjunctiva.[26][40] Classes of bacteria include Gram-positive cocci (e.g., Staphylococcus and Streptococcus) and Gram-negative rods and cocci (e.g., Haemophilus and Neisseria) are present.[40] Fungal genera include Candida, Aspergillus, and Penicillium.[26] The lachrymal glands continuously secrete, keeping the conjunctiva moist, while intermittent blinking lubricates the conjunctiva and washes away foreign material. Tears contain bactericides such as lysozyme, so that microorganisms have difficulty in surviving the lysozyme and settling on the epithelial surfaces.
Gut
The gut flora has the largest numbers of bacteria and the greatest number of species compared to other areas of the body.[6] In humans the gut flora is established at one to two years after birth, and by that time the intestinal epithelium and the intestinal mucosal barrier that it secretes have co-developed in a way that is tolerant to, and even supportive of, the gut flora and that also provides a barrier to pathogenic organisms.[19][45]
The relationship between some gut flora and humans is not merely commensal (a non-harmful coexistence), but rather a mutualistic relationship.[1] Some human gut microorganisms benefit the host by fermentating dietary fiber into short-chain fatty acids (SCFAs), such as acetic acid and butyric acid, which are then absorbed by the host.[6][46] Intestinal bacteria also play a role in synthesizing vitamin B and vitamin K as well as metabolizing bile acids, sterols, and xenobiotics.[1][46] The systemic importance of the SCFAs and other compounds they produce are like hormones and the gut flora itself appears to function like an endocrine organ,[46] and dysregulation of the gut flora has been correlated with a host of inflammatory and autoimmune conditions.[6][47]
The composition of human gut flora changes over time, when the diet changes, and as overall health changes.[6][47] A systematic review of 15 human randomized controlled trials from July 2016 found that certain commercially available strains of probiotic bacteria from the Bifidobacterium and Lactobacillus genera (B. longum, B. breve, B. infantis, L. helveticus, L. rhamnosus, L. plantarum, and L. casei), when taken by mouth in daily doses of 109–1010 colony forming units (CFU) for 1–2 months, possess treatment efficacy (i.e., improved behavioral outcomes) in certain central nervous system disorders – including anxiety, depression, autism spectrum disorder, and obsessive–compulsive disorder – and improved certain aspects of memory.[48]
Vagina
Vaginal microbiota refers to those species and genera that colonize the lower reproductive tract of women. These organisms play an important role in protecting against infections and maintaining vaginal health.[49] The most abundant vaginal microorganisms found in premenopausal women are from the genus Lactobacillus, which suppress pathogens by producing hydrogen peroxide and lactic acid.[28][49][50] Bacterial species composition and ratios vary depending on the stage of the menstrual cycle.[51][52] Ethnicity also influences vaginal flora. The occurrence of hydrogen peroxide-producing lactobacilli is lower in African American women and vaginal pH is higher.[53] Other influential factors such as sexual intercourse and antibiotics have been linked to the loss of lactobacilli.[50] Moreover, studies have found that sexual intercourse with a condom does appear to change lactobacilli levels, and does increase the level of Escherichia coli within the vaginal flora.[50] Changes in the normal, healthy vaginal microbiota is an indication of infections, such as candidiasis or bacterial vaginosis.[50] Candida albicans inhibits the growth of Lactobacillus species, while Lactobacillus species which produce hydrogen peroxide inhibit the growth and virulence of Candida albicans in both the vagina and the gut.[26][28][29]
Fungal genera that have been detected in the vagina include Candida, Pichia, Eurotium, Alternaria, Rhodotorula, and Cladosporium, among others.[26]
Placenta
Until recently the placenta was considered to be a sterile organ but commensal, nonpathogenic bacterial species and genera have been identified that reside in the placental tissue.[54][55][56]
Uterus
Until recently, the upper reproductive tract of women was considered to be a sterile environment. A variety of microorganisms inhabit the uterus of healthy, asymptomatic women of reproductive age. The microbiome of the uterus differs significantly from that of the vagina and gastrointestinal tract.[57]
Oral cavity
The environment present in the human mouth allows the growth of characteristic microorganisms found there. It provides a source of water and nutrients, as well as a moderate temperature.[1] Resident microbes of the mouth adhere to the teeth and gums to resist mechanical flushing from the mouth to stomach where acid-sensitive microbes are destroyed by hydrochloric acid.[1][28]
Anaerobic bacteria in the oral cavity include: Actinomyces, Arachnia, Bacteroides, Bifidobacterium, Eubacterium, Fusobacterium, Lactobacillus, Leptotrichia, Peptococcus, Peptostreptococcus, Propionibacterium, Selenomonas, Treponema, and Veillonella.[58] Genera of fungi that are frequently found in the mouth include Candida, Cladosporium, Aspergillus, Fusarium, Glomus, Alternaria, Penicillium, and Cryptococcus, among others.[26] Bacteria accumulate on both the hard and soft oral tissues in biofilms. Bacterial adhesion is particularly important for oral bacteria.
Oral bacteria have evolved mechanisms to sense their environment and evade or modify the host. Bacteria occupy the ecological niche provided by both the tooth surface and gingival epithelium. However, a highly efficient innate host defense system constantly monitors the bacterial colonization and prevents bacterial invasion of local tissues. A dynamic equilibrium exists between dental plaque bacteria and the innate host defense system.[59] Of particular interest is the role of oral microorganisms in the two major dental diseases: dental caries and periodontal disease.[59] Additionally, research has correlated poor oral heath and the resulting ability of the oral microbiota to invade the body to affect cardiac health as well as cognitive function.[60]
Lung
Much like the oral cavity, the upper and lower respiratory system possess mechanical deterrents to remove microbes. Goblet cells produce mucous which traps microbes and moves them out of the respiratory system via continuously moving ciliated epithelial cells.[1] In addition, a bactericidal effect is generated by nasal mucus which contains the enzyme lysozyme.[1]
Nonetheless, the upper and lower respiratory tract appears to have their own set of microbiota. Pulmonary bacterial microbiota belong to 9 major bacterial genera: Prevotella, Sphingomonas, Pseudomonas, Acinetobacter, Fusobacterium, Megasphaera, Veillonella, Staphylococcus, and Streptococcus. Some of the bacteria considered "normal biota" in the respiratory tract can cause serious disease especially in immunocompromised individuals; these include Streptococcus pyogenes, Haemophilus influenzae, Streptococcus pneumoniae, Neisseria meningitidis, and Staphylococcus aureus.
Fungal genera that compose the pulmonary mycobiome include Candida, Malassezia, Neosartorya, Saccharomyces, and Aspergillus, among others.[26]
Unusual distributions of bacterial and fungal genera in the respiratory tract is observed in people with cystic fibrosis.[26][61] Their bacterial flora often contains antibiotic-resistant and slow-growing bacteria, and the frequency of these pathogens changes in relation to age.[61]
Disease
Communities of microflora have been shown to change their behavior in diseased individuals.[62]
Cancer
Although cancer is generally a disease of host genetics and environmental factors, microorganisms are implicated in ~20% of human malignancies. Mucosal microbes can become part of the tumor microenvironment (TME) of aerodigestive tract malignancies. Intratumoral microbes can affect cancer growth and spread. Gut microbiota also detoxify dietary components, reducing inflammation and balancing host cell growth and proliferation. Coley's toxins were one of the earliest forms of cancer bacteriotherapy. Synthetic biology employs designer microbes and microbiota transplants against tumors.[63]
Microbes and the microbiota affect carcinogenesis in three broad ways: (i) altering the balance of tumor cell proliferation and death, (ii) regulating immune system function and (iii) influencing metabolism of host-produced factors, foods and pharmaceuticals.[63]
Modes of action
Ten microbes are designated by the International Agency for Research on Cancer (IARC) as human carcinogens. Most of these microbes colonize large percentages of the human population, although only genetically susceptible individuals develop cancer. Tumors arising at boundary surfaces, such as the skin, oropharynx and respiratory, digestive and urogenital tracts, harbor a microbiota, which complicates cancer-microbe causality. Substantial microbe presence at a tumor site does not establish association or causal links. Instead, microbes may find the tumor's oxygen tension or nutrient profile supportive. Decreased populations of specific microbes may also increase risks.[63]
Human oncoviruses can drive carcinogenesis by integrating oncogenes into host genomes. Human papillomaviruses (HPV) express oncoproteins such as E6 and E7. Viral integration selectively amplifies host genes in pathways with established cancer roles.[63]
Microbes affect genomic stability, resistance to cell death and proliferative signaling. Many bacteria can damage DNA, to kill competitors/survive. These defense factors can lead to mutational events that contribute to carcinogenesis. Examples include colibactin encoded by the pks locus (expressed by B2 group Escherichia coli as well as by other Enterobacteriaceae), Bacteroides fragilis toxin (Bft) produced by enterotoxigenic B. fragilis and cytolethal distending toxin (CDT) produced by several ε- and γ-proteobacteria. Colibactin is of interest in colorectal carcinogenesis, given the detection of pks+ E. coli in human colorectal cancers and the ability of colibactin-expressing E. coli to potentiate intestinal tumorigenesis in mice. Data also support a role for enterotoxigenic B. fragilis in both human and animal models of colon tumors. Both colibactin and CDT can cause double-stranded DNA damage in mammalian cells. In contrast, Bft acts indirectly by eliciting high levels of reactive oxygen species (ROS), which in turn damage host DNA. Chronically high ROS levels can outpace DNA repair mechanisms, leading to DNA damage and mutations.[63]
β-catenin
Several microbes possess proteins that engage host pathways involved in carcinogenesis. The Wnt/β-catenin signaling pathway, which regulates cells' polarity, growth and differentiation, is one example and is altered in many malignancies. Multiple cancer-associated bacteria can influence β-catenin signaling. Oncogenic type 1 strains of Helicobacter pylori express CagA, which is injected directly into the cytoplasm of host cells and modulates β-catenin to drive gastric cancer. This modulation leads to up-regulation of cellular proliferation, survival and migration genes, as well as angiogenesis—all processes central to carcinogenesis. Oral microbiota Fusobacterium nucleatum is associated with human colorectal adenomas and adenocarcinomas and amplified intestinal tumorigenesis in mice. F. nucleatum expresses FadA, a bacterial cell surface adhesion component that binds host E-cadherin, activating β-catenin. Enterotoxigenic B. fragilis, which is enriched in some human colorectal cancers, can stimulate E-cadherin cleavage via Btf, leading to β-catenin activation. Salmonella Typhi strains that maintain chronic infections secrete AvrA, which can activate epithelial β-catenin signaling and are associated with hepatobiliary cancers.[63]
Several of these bacteria are normal microbiota constituents. The presence of these cancer-potentiating microbes and their access to E-cadherin in evolving tumors demonstrate that a loss of appropriate boundaries and barrier maintenance between host and microbe is a critical step in the development of some tumors.[63]
Inflammation
Mucosal surface barriers are subject to environmental insult and must rapidly repair to maintain homeostasis. Compromised host or microbiota resiliency also reduce resistance to malignancy. Cancer and inflammatory disorders can then arise. Once barriers are breached, microbes can elicit proinflammatory or immunosuppressive programs.[63]
Inflammation, whether high-grade as in inflammatory disorders or low-grade as in malignancies and obesity, drive a tumor-permissive milieu. Pro-inflammatory factors such as reactive oxygen and nitrogen species, cytokines and chemokines can also drive tumor growth and spread. Tumors can up-regulate and activate pattern recognition receptors (e.g. toll-like receptors), driving feedforward loops of activation of cancer-associated inflammation regulator NF-κΒ. Cancer-associated microbes appear to activate NF-κΒ signaling within the TME. The activation of NF-κΒ by F. nucleatum may be the result of pattern recognition receptor engagement or FadA engagement of E-cadherin. Other pattern recognition receptors, such as nucleotide-binding oligomerization domain–like receptor (NLR) family members NOD-2, NLRP3, NLRP6 and NLRP12, may play a role in mediating colorectal cancer.[63]
Immune system TME engagement is not restricted to the innate immune system. Once the innate immune system is activated, adaptive immune responses ensue, often with tumor progression. The interleukin-23 (IL-23)–IL-17 axis, tumor necrosis factor–α (TNF-α)–TNF receptor signaling, IL-6–IL-6 family member signaling, and STAT3 activation all represent innate and adaptive pathways contributing to tumor progression and growth.[63]
The microbiota adapts to host changes such as inflammation. Adaptation shift microbiota to a vulnerable tissue site. Genotoxin azoxymethane and colon barrier–disrupting agent dextran sodium sulfate independently result in colon tumors in susceptible mouse strains; combining them accelerates tumorigenesis.[63]
Perturbations to a host immune system coupled with inflammatory stimulus may enrich bacterial clades that attach to host surfaces, invade host tissue, or trigger host inflammatory mediators. Fecal microbiota from NOD2- or NLRP6-deficient mice acquire features that enhance the susceptibility of wild-type mice to caCRC. In mice, gut microbiota modulate colon tumorigenesis, independent of genetic deficiencies. Germ-free mice developed more tumors when colonized from donors with caCRC, once followed by treatments that induced caCRC.[63]
Research
Diabetic foot ulcers develop their own, distinctive microbiota. Investigations into characterizing and identifying the phyla, genera and species of bacteria or other microorganisms populating these ulcers may help identify one group of microbiota that promotes healing.[64]
Environmental health
Studies in 2009 questioned whether the decline in biota (including microfauna) as a result of human intervention might impede human health.[65]
See also
References
- 1 2 3 4 5 6 7 8 9 10 11 Sherwood, Linda; Willey, Joanne; Woolverton, Christopher (2013). Prescott's Microbiology (9th ed.). New York: McGraw Hill. pp. 713–721. ISBN 9780073402406. OCLC 886600661.
- 1 2 3 American Academy of Microbiology FAQ: Human Microbiome January 2014
- 1 2 Judah L. Rosner for Microbe Magazine, Feb 2014. Ten Times More Microbial Cells than Body Cells in Humans?
- 1 2 Alison Abbott for Nature News. Jan 8 2016 Scientists bust myth that our bodies have more bacteria than human cells
- 1 2 Sender, R; Fuchs, S; Milo, R (Jan 2016). "Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans". Cell. 164 (3): 337–40. doi:10.1016/j.cell.2016.01.013. PMID 26824647.
- 1 2 3 4 5 6 7 Quigley, EM (2013). "Gut bacteria in health and disease". Gastroenterol Hepatol (N Y). 9 (9): 560–9. PMC 3983973
 . PMID 24729765.
. PMID 24729765. - ↑ Falony G, Vieira-Silva S, Raes J (2015). "Microbiology Meets Big Data: The Case of Gut Microbiota-Derived Trimethylamine". Annu. Rev. Microbiol. 69: 305–321. doi:10.1146/annurev-micro-091014-104422. PMID 26274026.
we review literature on trimethylamine (TMA), a microbiota-generated metabolite linked to atherosclerosis development.
- ↑ Gaci N, Borrel G, Tottey W, O'Toole PW, Brugère JF (November 2014). "Archaea and the human gut: new beginning of an old story". World J. Gastroenterol. 20 (43): 16062–16078. doi:10.3748/wjg.v20.i43.16062. PMC 4239492
 . PMID 25473158.
. PMID 25473158. Trimethylamine is exclusively a microbiota-derived product of nutrients (lecithin, choline, TMAO, L-carnitine) from normal diet, from which seems originate two diseases, trimethylaminuria (or Fish-Odor Syndrome) and cardiovascular disease through the proatherogenic property of its oxidized liver-derived form.
- 1 2 "NIH Human Microbiome Project defines normal bacterial makeup of the body". NIH News. 13 June 2012.
- 1 2 3 NIH Human Microbiome Working Group (2009). "The NIH Human Microbiome Project". Genome Res. 19 (12): 2317–2323. doi:10.1101/gr.096651.109. PMC 2792171
 . PMID 19819907.
. PMID 19819907. - ↑ Kuczynski, J.; et al. (2011). "Experimental and analytical tools for studying the human microbiome". Nature Reviews Genetics. 13 (1): 47–58. doi:10.1038/nrg3129. PMID 22179717.
- ↑ Vestheim, H.; Jarman, S. N. (2008). "Blocking primers to enhance PCR amplification of rare sequences in mixed samples – a case study on prey DNA in Antarctic krill stomachs". Frontiers in Zoology. 5: 12. doi:10.1186/1742-9994-5-12. PMC 2517594
 . PMID 18638418.
. PMID 18638418. - ↑ Tap, Julien; Mondot; et al. (2009). "Towards the human intestinal microbiota phylogenetic core". Environmental Microbiology. 11 (102): 2574–2584. doi:10.1111/j.1462-2920.2009.01982.x. PMID 19601958.
- ↑ Hamady, M.; Knight, R. (2009). "Microbial community profiling for human microbiome projects: Tools, techniques, and challenges". Genome Research. 19 (7): 1141–1152. doi:10.1101/gr.085464.108. PMID 19383763.
- ↑ Human Microbiome Project Consortium (2012). "A framework for human microbiome research". Nature. 486 (7402): 215–221. Bibcode:2012Natur.486..215T. doi:10.1038/nature11209. PMC 3377744
 . PMID 22699610.
. PMID 22699610. - ↑ The Human Microbiome Project Consortium (2012). "Structure, function and diversity of the healthy human microbiome". Nature. 486 (7402): 207–214. Bibcode:2012Natur.486..207T. doi:10.1038/nature11234. PMC 3564958
 . PMID 22699609.
. PMID 22699609. - ↑ PLoS Human Microbiome Project Collection Manuscript Summaries June 13, 2012
- ↑ "Consortium of Scientists Map the Human Body's Bacterial Ecosystem". ucsf.edu.
- 1 2 Sommer, F; Bäckhed, F (Apr 2013). "The gut microbiota--masters of host development and physiology". Nat Rev Microbiol. 11 (4): 227–38. doi:10.1038/nrmicro2974. PMID 23435359.
- ↑ Eckburg PB, Bik EM, Bernstein CN, et al. (2005). "Diversity of the human intestinal microbial flora". Science. 308 (5728): 1635–8. Bibcode:2005Sci...308.1635E. doi:10.1126/science.1110591. PMC 1395357
 . PMID 15831718.
. PMID 15831718. - ↑ Duncan SH, Louis P, Flint HJ (2007). "Cultivable bacterial diversity from the human colon". Lett. Appl. Microbiol. 44 (4): 343–50. doi:10.1111/j.1472-765X.2007.02129.x. PMID 17397470.
- ↑ Florin TH, Zhu G, Kirk KM, Martin NG (2000). "Shared and unique environmental factors determine the ecology of methanogens in humans and rats". Am. J. Gastroenterol. 95 (10): 2872–9. doi:10.1111/j.1572-0241.2000.02319.x. PMID 11051362.
- ↑ Eckburg P, Lepp P, Relman D (2003). "Archaea and their potential role in human disease". Infection and Immunity. 71 (2): 591–6. doi:10.1128/IAI.71.2.591-596.2003. PMC 145348
 . PMID 12540534.
. PMID 12540534. - ↑ Cavicchioli R, Curmi P, Saunders N, Thomas T (2003). "Pathogenic archaea: do they exist?". BioEssays. 25 (11): 1119–28. doi:10.1002/bies.10354. PMID 14579252.
- ↑ Lepp P, Brinig M, Ouverney C, Palm K, Armitage G, Relman D (2004). "Methanogenic Archaea and human periodontal disease". Proc. Natl. Acad. Sci. U.S.A. 101 (16): 6176–81. Bibcode:2004PNAS..101.6176L. doi:10.1073/pnas.0308766101. PMC 395942
 . PMID 15067114.
. PMID 15067114. - 1 2 3 4 5 6 7 8 9 10 11 Cui L, Morris A, Ghedin E (July 2013). "The human mycobiome in health and disease". Genome Med. 5 (7): 63. doi:10.1186/gm467. PMC 3978422
 . PMID 23899327.
. PMID 23899327. Figure 2: Distribution of fungal genera in different body sites
- 1 2 Martins N, Ferreira IC, Barros L, Silva S, Henriques M (June 2014). "Candidiasis: predisposing factors, prevention, diagnosis and alternative treatment". Mycopathologia. 177 (5–6): 223–240. doi:10.1007/s11046-014-9749-1. PMID 24789109.
Candida species and other microorganisms are involved in this complicated fungal infection, but Candida albicans continues to be the most prevalent. In the past two decades, it has been observed an abnormal overgrowth in the gastrointestinal, urinary and respiratory tracts, not only in immunocompromised patients, but also related to nosocomial infections and even in healthy individuals. There is a widely variety of causal factors that contribute to yeast infection which means that candidiasis is a good example of a multifactorial syndrome.
- 1 2 3 4 5 Wang ZK, Yang YS, Stefka AT, Sun G, Peng LH (April 2014). "Review article: fungal microbiota and digestive diseases". Aliment. Pharmacol. Ther. 39 (8): 751–766. doi:10.1111/apt.12665. PMID 24612332.
In addition, GI fungal infection is reported even among those patients with normal immune status. Digestive system-related fungal infections may be induced by both commensal opportunistic fungi and exogenous pathogenic fungi. ... Candida sp. is also the most frequently identified species among patients with gastric IFI. ... It was once believed that gastric acid could kill microbes entering the stomach and that the unique ecological environment of the stomach was not suitable for microbial colonisation or infection. However, several studies using culture-independent methods confirmed that large numbers of acid-resistant bacteria belonging to eight phyla and up to 120 species exist in the stomach, such as Streptococcus sp., Neisseria sp. and Lactobacillus sp. etc.26, 27 Furthermore, Candida albicans can grow well in highly acidic environments,28 and some genotypes may increase the severity of gastric mucosal lesions.29
- 1 2 3 Erdogan A, Rao SS (April 2015). "Small intestinal fungal overgrowth". Curr Gastroenterol Rep. 17 (4): 16. doi:10.1007/s11894-015-0436-2. PMID 25786900.
Small intestinal fungal overgrowth (SIFO) is characterized by the presence of excessive number of fungal organisms in the small intestine associated with gastrointestinal (GI) symptoms. Candidiasis is known to cause GI symptoms particularly in immunocompromised patients or those receiving steroids or antibiotics. However, only recently, there is emerging literature that an overgrowth of fungus in the small intestine of non-immunocompromised subjects may cause unexplained GI symptoms. Two recent studies showed that 26 % (24/94) and 25.3 % (38/150) of a series of patients with unexplained GI symptoms had SIFO. The most common symptoms observed in these patients were belching, bloating, indigestion, nausea, diarrhea, and gas. ... Fungal-bacterial interaction may act in different ways and may either be synergistic or antagonistic or symbiotic [29]. Some bacteria such as Lactobacillus species can interact and inhibit both the virulence and growth of Candida species in the gut by producing hydrogen peroxide [30]. Any damage to the mucosal barrier or disruption of GI microbiota with chemotherapy or antibiotic use, inflammatory processes, activation of immune molecules and disruption of epithelial repair may all cause fungal overgrowth [27].
- ↑ Marcon MJ, Powell DA (1 April 1992). "Human infections due to Malassezia spp". Clin. Microbiol. Rev. 5 (2): 101–19. PMC 358230
 . PMID 1576583.
. PMID 1576583. - ↑ Roth RR, James WD (1988). "Microbial ecology of the skin". Annu. Rev. Microbiol. 42 (1): 441–64. doi:10.1146/annurev.mi.42.100188.002301. PMID 3144238.
- ↑ Hannigan, Geoffrey D.; Meisel, Jacquelyn S.; Tyldsley, Amanda S.; Zheng, Qi; Hodkinson, Brendan P.; SanMiguel, Adam J.; Minot, Samuel; Bushman, Frederic D.; Grice, Elizabeth A. (2015-10-30). "The Human Skin Double-Stranded DNA Virome: Topographical and Temporal Diversity, Genetic Enrichment, and Dynamic Associations with the Host Microbiome". MBio. 6 (5): e01578–15. doi:10.1128/mBio.01578-15. ISSN 2150-7511. PMC 4620475
 . PMID 26489866.
. PMID 26489866. - ↑ Minot, Samuel; Sinha, Rohini; Chen, Jun; Li, Hongzhe; Keilbaugh, Sue A.; Wu, Gary D.; Lewis, James D.; Bushman, Frederic D. (2011-10-01). "The human gut virome: inter-individual variation and dynamic response to diet". Genome Research. 21 (10): 1616–1625. doi:10.1101/gr.122705.111. ISSN 1549-5469. PMC 3202279
 . PMID 21880779.
. PMID 21880779. - ↑ Young, J. C.; Chehoud, C.; Bittinger, K.; Bailey, A.; Diamond, J. M.; Cantu, E.; Haas, A. R.; Abbas, A.; Frye, L. (2015-01-01). "Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients". American Journal of Transplantation: Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 15 (1): 200–209. doi:10.1111/ajt.13031. ISSN 1600-6143. PMC 4276431
 . PMID 25403800.
. PMID 25403800. - ↑ Abeles, Shira R.; Robles-Sikisaka, Refugio; Ly, Melissa; Lum, Andrew G.; Salzman, Julia; Boehm, Tobias K.; Pride, David T. (2014-09-01). "Human oral viruses are personal, persistent and gender-consistent". The ISME Journal. 8 (9): 1753–1767. doi:10.1038/ismej.2014.31. ISSN 1751-7362. PMC 4139723
 . PMID 24646696.
. PMID 24646696. - ↑ Ly, Melissa; Abeles, Shira R.; Boehm, Tobias K.; Robles-Sikisaka, Refugio; Naidu, Mayuri; Santiago-Rodriguez, Tasha; Pride, David T. (2014-07-01). "Altered Oral Viral Ecology in Association with Periodontal Disease". MBio. 5 (3): e01133–14. doi:10.1128/mBio.01133-14. ISSN 2150-7511. PMC 4030452
 . PMID 24846382.
. PMID 24846382. - ↑ Monaco, Cynthia L.; Gootenberg, David B.; Zhao, Guoyan; Handley, Scott A.; Ghebremichael, Musie S.; Lim, Efrem S.; Lankowski, Alex; Baldridge, Megan T.; Wilen, Craig B. (2016-03-09). "Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome". Cell Host & Microbe. 19 (3): 311–322. doi:10.1016/j.chom.2016.02.011. ISSN 1934-6069. PMC 4821831
 . PMID 26962942.
. PMID 26962942. - ↑ Norman, Jason M.; Handley, Scott A.; Baldridge, Megan T.; Droit, Lindsay; Liu, Catherine Y.; Keller, Brian C.; Kambal, Amal; Monaco, Cynthia L.; Zhao, Guoyan (2015-01-29). "Disease-specific alterations in the enteric virome in inflammatory bowel disease". Cell. 160 (3): 447–460. doi:10.1016/j.cell.2015.01.002. ISSN 1097-4172. PMC 4312520
 . PMID 25619688.
. PMID 25619688. - ↑ Grice, Elizabeth A.; Kong, HH; Conlan, S; Deming, CB; Davis, J; Young, AC; NISC Comparative Sequencing Program; Bouffard, GG; et al. (2006). "Topographical and Temporal Diversity of the Human Skin Microbiome". Science. 324 (5931): 1190–2. Bibcode:2009Sci...324.1190G. doi:10.1126/science.1171700. PMC 2805064
 . PMID 19478181.
. PMID 19478181. - 1 2 "The Normal Bacterial Flora of Humans". textbookofbacteriology.net.
- 1 2 3 4 5 6 7 8 9 Zhang LS, Davies SS (April 2016). "Microbial metabolism of dietary components to bioactive metabolites: opportunities for new therapeutic interventions". Genome Med. 8 (1): 46. doi:10.1186/s13073-016-0296-x. PMC 4840492
 . PMID 27102537.
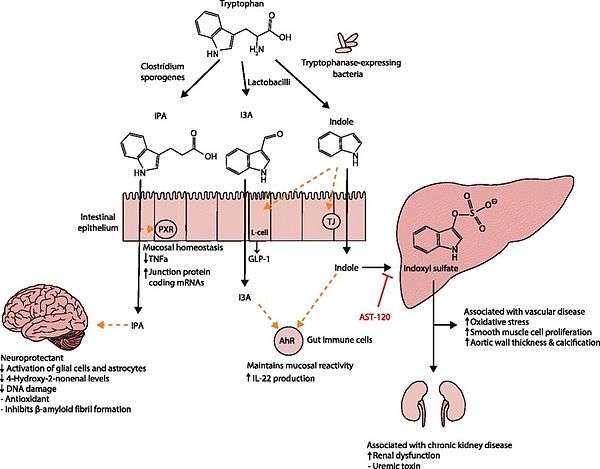
. PMID 27102537. Lactobacillus spp. convert tryptophan to indole-3-aldehyde (I3A) through unidentified enzymes [125]. Clostridium sporogenes convert tryptophan to IPA [6], likely via a tryptophan deaminase. ... IPA also potently scavenges hydroxyl radicals
Table 2: Microbial metabolites: their synthesis, mechanisms of action, and effects on health and disease
Figure 1: Molecular mechanisms of action of indole and its metabolites on host physiology and disease - ↑ Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G (2009). "Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites". Proc. Natl. Acad. Sci. U.S.A. 106 (10): 3698–703. doi:10.1073/pnas.0812874106. PMC 2656143
 . PMID 19234110.
. PMID 19234110. Production of IPA was shown to be completely dependent on the presence of gut microflora and could be established by colonization with the bacterium Clostridium sporogenes.
IPA metabolism diagram - ↑ "3-Indolepropionic acid". Human Metabolome Database. University of Alberta. Retrieved 12 October 2015.
Indole-3-propionate (IPA), a deamination product of tryptophan formed by symbiotic bacteria in the gastrointestinal tract of mammals and birds. 3-Indolepropionic acid has been shown to prevent oxidative stress and death of primary neurons and neuroblastoma cells exposed to the amyloid beta-protein in the form of amyloid fibrils, one of the most prominent neuropathologic features of Alzheimer's disease. 3-Indolepropionic acid also shows a strong level of neuroprotection in two other paradigms of oxidative stress. (PMID 10419516 )
Origin: • Endogenous • Microbial - ↑ Chyan YJ, Poeggeler B, Omar RA, Chain DG, Frangione B, Ghiso J, Pappolla MA (1999). "Potent neuroprotective properties against the Alzheimer beta-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid". J. Biol. Chem. 274 (31): 21937–42. doi:10.1074/jbc.274.31.21937. PMID 10419516.
[Indole-3-propionic acid (IPA)] has previously been identified in the plasma and cerebrospinal fluid of humans, but its functions are not known. ... In kinetic competition experiments using free radical-trapping agents, the capacity of IPA to scavenge hydroxyl radicals exceeded that of melatonin, an indoleamine considered to be the most potent naturally occurring scavenger of free radicals. In contrast with other antioxidants, IPA was not converted to reactive intermediates with pro-oxidant activity.
- ↑ Faderl, M; et al. (Apr 2015). "Keeping bugs in check: The mucus layer as a critical component in maintaining intestinal homeostasis". IUBMB Life. 67 (4): 275–85. doi:10.1002/iub.1374. PMID 25914114.
- 1 2 3 Clarke, G; et al. (Aug 2014). "Minireview: Gut microbiota: the neglected endocrine organ". Mol Endocrinol. 28 (8): 1221–38. doi:10.1210/me.2014-1108. PMID 24892638.
- 1 2 Shen S, Wong CH. "Bugging inflammation: role of the gut microbiota. Clin Transl Immunology. 2016 Apr;5(4):e72. Review. doi:10.1038/cti.2016.12 PMID 27195115 PDF
- ↑ Wang H, Lee IS, Braun C, Enck P (July 2016). "Effect of probiotics on central nervous system functions in animals and humans - a systematic review". J. Neurogastroenterol Motil. doi:10.5056/jnm16018. PMID 27413138.
These probiotics showed efficacy in improving psychiatric disorder-related behaviors including anxiety, depression, autism spectrum disorder (ASD), obsessive-compulsive disorder, and memory abilities, including spatial and non-spatial memory. Because many of the basic science studies showed some efficacy of probiotics on central nervous system function, this background may guide and promote further preclinical and clinical studies. ... According to the qualitative analyses of current studies, we can provisionally draw the conclusion that B. longum, B. breve, B. infantis, L. helveticus, L. rhamnosus, L. plantarum, and L. casei were most effective in improving CNS function, including psychiatric disease-associated functions (anxiety, depression, mood, stress response) and memory abilities
- 1 2 Petrova, Mariya I.; Lievens, Elke; Malik, Shweta; Imholz, Nicole; Lebeer, Sarah (2015). "Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health". Frontiers in Physiology. 6. doi:10.3389/fphys.2015.00081. ISSN 1664-042X.
- 1 2 3 4 Witkin, S. S.; Linhares, I. M.; Giraldo, P. (2007). "Bacterial flora of the female genital tract: Function and immune regulation". Best Practice & Research Clinical Obstetrics & Gynaecology. 21 (3): 347–354. doi:10.1016/j.bpobgyn.2006.12.004. PMID 17215167.
- ↑ Todar, Kenneth (2012). "The Normal Bacterial Flora of Humans". Todar's Online Textbook of Bacteriology. Madison, WI: Kenneth Todar. Retrieved 2012-04-06.
- ↑ Onderdonk, A. B.; Zamarchi, G. R.; Walsh, J. A.; Mellor, R. D.; Muñoz, A.; Kass, E. H. (1986). "Methods for quantitative and qualitative evaluation of vaginal microflora during menstruation". Applied and Environmental Microbiology. 51 (2): 333–339. PMC 238869
 . PMID 3954346.
. PMID 3954346. - ↑ Antonio, M. A. D.; Hawes, S. E.; Hillier, S. L. (1999). "The Identification of VaginalLactobacillusSpecies and the Demographic and Microbiologic Characteristics of Women Colonized by These Species". The Journal of Infectious Diseases. 180 (6): 1950–1956. doi:10.1086/315109. PMID 10558952.
- ↑ Fox, Chelsea; Eichelberger, Kacey (2015). "Maternal microbiome and pregnancy outcomes". Fertility and Sterility. 104 (6): 1358–1363. doi:10.1016/j.fertnstert.2015.09.037. ISSN 0015-0282. PMID 26493119.
- ↑ Wassenaar, T.M.; Panigrahi, P. (2014). "Is a foetus developing in a sterile environment?". Letters in Applied Microbiology. 59 (6): 572–579. doi:10.1111/lam.12334. ISSN 0266-8254. PMID 25273890.
- ↑ Schwiertz, Andreas (2016). Microbiota of the human body : implications in health and disease. Switzerland: Springer. p. 1. ISBN 978-3-319-31248-4.
- ↑ Franasiak, Jason M.; Scott, Richard T. (2015). "Reproductive tract microbiome in assisted reproductive technologies". Fertility and Sterility. 104 (6): 1364–1371. doi:10.1016/j.fertnstert.2015.10.012. ISSN 0015-0282. PMID 26597628.
- ↑ Sutter, V. L. (1984). "Anaerobes as normal oral flora". Reviews of infectious diseases. 6 Suppl 1: S62–S66. doi:10.1093/clinids/6.Supplement_1.S62. PMID 6372039.
- 1 2 Rogers A H (editor). (2008). Molecular Oral Microbiology. Caister Academic Press. ISBN 978-1-904455-24-0.
- ↑ Noble JM, Scarmeas N, Papapanou PN (2013). "Poor oral health as a chronic, potentially modifiable dementia risk factor: review of the literature.". Curr Neurol Neurosci Rep. 13 (10): 384. doi:10.1007/s11910-013-0384-x. PMID 23963608.
- 1 2 Beringer, P M; M D Appleman (November 2000). "Unusual respiratory bacterial flora in cystic fibrosis: microbiologic and clinical features" (PDF). Current Opinion in Pulmonary Medicine. 6 (6): 545–550. doi:10.1097/00063198-200011000-00015. ISSN 1070-5287. PMID 11100967.
- ↑ "Mouth bacteria can change its diet, supercomputers reveal". medicalxpress.com.
- 1 2 3 4 5 6 7 8 9 10 11 12 Garrett, Wendy S. (3 April 2015). "Cancer and the microbiota". Science Magazine. doi:10.1126/science.aaa4972. Retrieved 2015-06-29.
- ↑ Lavigne, Jean-Philippe; Sotto, Albert; Dunyach-Remy, Catherine; Lipsky, Benjamin A. (2015). "New Molecular Techniques to Study the Skin Microbiota of Diabetic Foot Ulcers". Advances in Wound Care. 4 (1): 38–49. doi:10.1089/wound.2014.0532. ISSN 2162-1918. PMC 4281861
 . PMID 25566413.
. PMID 25566413. - ↑ Katherine Harmon (16 December 2009). "Bugs Inside: What Happens When the Microbes That Keep Us Healthy Disappear?". Scientific American. Retrieved 27 December 2008.
External links
- The Secret World Inside You Exhibit 2015-2016, American Museum of Natural History
- FAQ: Human Microbiome, January 2014 American Society For Microbiology