Bacillus subtilis
| Bacillus subtilis | |
|---|---|
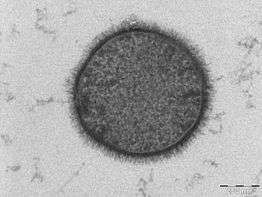 | |
| TEM micrograph of a B. subtilis cell in cross-section (scale bar = 200 nm) | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Firmicutes |
| Class: | Bacilli |
| Order: | Bacillales |
| Family: | Bacillaceae |
| Genus: | Bacillus |
| Species: | B. subtilis |
| Binomial name | |
| Bacillus subtilis (Ehrenberg 1835) Cohn 1872 | |
| Synonyms | |
| |
Bacillus subtilis, known also as the hay bacillus or grass bacillus, is a Gram-positive, catalase-positive bacterium, found in soil and the gastrointestinal tract of ruminants and humans. A member of the genus Bacillus, B. subtilis is rod-shaped, and can form a tough, protective endospore, allowing it to tolerate extreme environmental conditions. B. subtilis has historically been classified as an obligate aerobe, though evidence exists that it is a facultative aerobe. B. subtilis is considered the best studied Gram-positive bacterium and a model organism to study bacterial chromosome replication and cell differentiation. It is one of the bacterial champions in secreted enzyme production and used on an industrial scale by biotechnology companies.
Description
Bacillus subtilis is a Gram-positive bacterium, rod-shaped and catalase-positive. It was originally named Vibrio subtilis by Christian Gottfried Ehrenberg,[3] and renamed Bacillus subtilis by Ferdinand Cohn in 1872[4] (subtilis being the Latin for 'fine'). B. subtilis cells are typically rod-shaped, and are about 4-10 micrometers (μm) long and 0.25–1.0 μm in diameter, with a cell volume of about 4.6 fL at stationary phase.[5] As with other members of the genus Bacillus, it can form an endospore, to survive extreme environmental conditions of temperature and desiccation.[6] B. subtilis is a facultative anaerobe[7] and had been considered as an obligate aerobe until 1998. B. subtilis is heavily flagellated, which gives it the ability to move quickly in liquids. B. subtilis has proven highly amenable to genetic manipulation, and has become widely adopted as a model organism for laboratory studies, especially of sporulation, which is a simplified example of cellular differentiation. In terms of popularity as a laboratory model organism, B. subtilis is often considered as the Gram-positive equivalent of Escherichia coli, an extensively studied Gram-negative bacterium.
Below is a table that helps identify B. subtilis through certain identification tests.
| Test | Result[8][9][10][11] |
|---|---|
| Acid from Glucose | + |
| Acid from Lactose | - |
| Acid from Sucrose | + |
| Acid from Mannitol | + |
| Catalase | + |
| Citrate (Simmons) | + |
| Endospore | + |
| Gas from Glucose | - |
| Gelatin Hydrolysis | + |
| Gram Stain | + |
| Hydrogen Sulfide Production | - |
| Indole Production | - |
| Motility | + |
| Nitrate Reduction | + |
| Oxidase | + |
| Phenylalanine Deaminase | - |
| Shape | Rod |
| Urea Hydrolysis | - |
| Voges-Proskaeur-Proskaeur | + |
Habitat
This species is commonly found in the upper layers of the soil, and evidence exists that B. subtilis is a normal gut commensal in humans. A 2009 study compared the density of spores found in soil (about 106 spores per gram) to that found in human feces (about 104 spores per gram). The number of spores found in the human gut was too high to be attributed solely to consumption through food contamination.[12]
Reproduction


B. subtilis can divide symmetrically to make two daughter cells (binary fission), or asymmetrically, producing a single endospore that can remain viable for decades and is resistant to unfavourable environmental conditions such as drought, salinity, extreme pH, radiation, and solvents. The endospore is formed at times of nutritional stress, allowing the organism to persist in the environment until conditions become favourable. Prior to the process of sporulation the cells might become motile by producing flagella, take up DNA from the environment, or produce antibiotics. These responses are viewed as attempts to seek out nutrients by seeking a more favourable environment, enabling the cell to make use of new beneficial genetic material or simply by killing of competition.
Under stressful conditions, such as nutrient deprivation, B. subtilis undergoes the process of sporulation to ensure the survival of the species. This process has been very well studied and has served as a model organism for studying sporulation.
Chromosomal replication
B. subtilis is a model organism used to study bacterial chromosome replication. Replication of the single circular chromosome initiates at a single locus, the origin (oriC). Replication proceeds bidirectionally and two replication forks progress in clockwise and counterclockwise directions along the chromosome. Chromosome replication is completed when the forks reach the terminus region, which is positioned opposite to the origin on the chromosome map. The terminus region contains several short DNA sequences (Ter sites) that promote replication arrest. Specific proteins mediate all the steps in DNA replication. Comparison between the proteins involved in chromosomal DNA replication in B. subtilis and in Escherichia coli reveals similarities and differences. Although the basic components promoting initiation, elongation, and termination of replication are well-conserved, some important differences can be found (such as one bacterium missing proteins essential in the other). These differences underline the diversity in the mechanisms and strategies that various bacterial species have adopted to carry out the duplication of their genomes.[13]
Genome
B. subtilis has about 4,100 genes. Of these, only 192 were shown to be indispensable; another 79 were predicted to be essential, as well. A vast majority of essential genes were categorized in relatively few domains of cell metabolism, with about half involved in information processing, one-fifth involved in the synthesis of cell envelope and the determination of cell shape and division, and one-tenth related to cell energetics.[14]
The complete genome sequence of B. subtilis sub-strain QB928 has 4,146,839 DNA base pairs and 4,292 genes. The QB928 strain is widely used in genetic studies due to the presence of various markers [aroI(aroK)906 purE1 dal(alrA)1 trpC2].[15]
Several noncoding RNAs have been characterized in the B. subtilis genome in 2009, including Bsr RNAs.[16] Microarray-based comparative genomic analyses have revealed that B. subtilis members show considerable genomic diversity.[17]
Transformation
Natural bacterial transformation involves the transfer of DNA from one bacterium to another through the surrounding medium. In B. subtilis, length of transferred DNA is greater than 1271 kb (more than 1 million bases).[18] The transferred DNA is likely double-stranded DNA and is often more than a third of the total chromosome length of 4215 kb.[19] It appears that about 7-9% of the recipient cells take up an entire chromosome.[20]
In order for a recipient bacterium to bind, take up exogenous DNA from another bacterium of the same species and recombine it into its chromosome, it must enter a special physiological state called competence. Competence in B. subtilis is induced toward the end of logarithmic growth, especially under conditions of amino-acid limitation.[21] Under these stressful conditions of semistarvation, cells typically have just one copy of their chromosome and likely have increased DNA damage. To test whether transformation is an adaptive function for B. subtilis to repair its DNA damage, experiments were conducted using UV light as the damaging agent.[22][23][24] These experiments led to the conclusion that competence, with uptake of DNA, is specifically induced by DNA-damaging conditions, and that transformation functions as a process for recombinational repair of DNA damage.[25]
Uses
1900s
Cultures of B. subtilis were popular worldwide before the introduction of antibiotics as an immunostimulatory agent to aid treatment of gastrointestinal and urinary tract diseases. It was used throughout the 1950s as an alternative medicine, which upon digestion has been found to significantly stimulate broad-spectrum immune activity including activation of secretion of specific antibodies IgM, IgG and IgA[26] and release of CpG dinucleotides inducing INF A/Y producing activity of leukocytes and cytokines important in the development of cytotoxicity towards tumor cells.[27] It was marketed throughout America and Europe from 1946 as an immunostimulatory aid in the treatment of gut and urinary tract diseases such as Rotavirus and Shigellosis,[28] but declined in popularity after the introduction of antibiotics, despite causing fewer allergic reactions and significantly lower toxicity to normal gut flora. It is still widely used in Western Europe and the Middle East as an alternative medicine.The high stability of B. subtilis in harsh environmental conditions makes this microorganism a perfect candidate for probiotics applications either in baked and pasteurized foods/beverages or in other galenic forms like tablets, capsules, and powder.
Since the 1960s B. subtilis has had a history as a test species in spaceflight experimentation. Its endospores can survive up to 6 years in space if coated by dust particles protecting it from solar UV rays.[29]* It has been used as an extremophile survival indicator in outer space such as Exobiology Radiation Assembly,[30][31] EXOSTACK,[32][33] and EXPOSE orbital missions.[34][35][36]
Wild-type natural isolates of B. subtilis are difficult to work with compared to laboratory strains that have undergone domestication processes of mutagenesis and selection. These strains often have improved capabilities of transformation (uptake and integration of environmental DNA), growth, and loss of abilities needed "in the wild". And, while dozens of different strains fitting this description exist, the strain designated '168' is the most widely used.

B. globigii, a closely related but phylogenetically distinct species now known as Bacillus atrophaeus[37][38] was used as a biowarfare simulant during Project SHAD (aka Project 112).[39] Subsequent genomic analysis showed that the strains used in those studies were products of deliberate enrichment for strains that exhibited abnormally high rates of sporulation.[40]
A strain of B. subtilis formerly known as Bacillus natto is used in the commercial production of the Japanese food natto, as well as the similar Korean food cheonggukjang.
2000s
- As a model organism, B. subtilis is commonly used in laboratory studies directed at discovering the fundamental properties and characteristics of Gram-positive spore-forming bacteria.[41] In particular, the basic principles and mechanisms underlying formation of the durable endospore have been deduced from studies of spore formation in B. subtilis.
- It can convert some explosives into harmless compounds of nitrogen, carbon dioxide, and water.
- Its surface-binding properties play a role in safe radionuclide waste [e.g. thorium (IV) and plutonium (IV)] disposal.
- Recombinant strains pBE2C1 and pBE2C1AB were used in production of polyhydroxyalkanoates (PHA), and malt waste can be used as their carbon source for lower-cost PHA production.
- Due to its excellent fermentation properties, with high product yields (20 to 25 gram per litre) it is used to produce various enzymes, such as amylase and proteases.[42]
- Other enzymes produced by B. subtilis and B. licheniformis are widely used as additives in laundry detergents.
- It is used to produce hyaluronic acid, which is used in the joint-care sector in healthcare[43] and cosmetics.
- B. subtilis is used as a soil inoculant in horticulture and agriculture.[44][45][46]
- It may provide some benefit to saffron growers by speeding corm growth and increasing stigma biomass yield.[47]
- Monsanto has isolated a gene from B. subtilis that expresses cold shock protein B and spliced it into their drought-tolerant corn hybrid MON 87460, which was approved for sale in the US in November 2011.[48][49]
- It is used as an "indicator organism" during gas sterilization procedures, to ensure a sterilization cycle has completed successfully.[50][51] This is due to the difficulty in sterilizing endospores.
- Novel strains of B. subtilis that could use 4-fluorotryptophan (4FTrp) but not canonical tryptophan (Trp) for propagation were isolated. As Trp is only coded by a single codon, there is evidence that Trp can be displaced by 4FTrp in the genetic code. The experiments showed that the canonical genetic code can be mutable.[52]
- Bacillus subtilis strain KPA has the ability to enhance industrially important enzymes, especially amylase under mild stress conditions.
- B. subtilis (natto) enhances the growth and (or) viability of lactobacilli, possibly through production of catalase and subtilisin.[53]
- Bacillus subtilis is currently being used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders (mostly as a direct result of antibiotic treatment), many of which lead to diarrhea. Ingestion of significant quantities of Bacillus subtilis is thought to restore the normal microbial flora following extensive antibiotic use or illness.[54]
- Bacillus subtilis containing pharmaceutical preparations has been widely used in the treatment and prophylaxis of intestinal disorders associated with antibiotic therapy or diarrhoea of various origin.[55]
- Treatment with Bacillus subtilis probiotics during antibiotic therapy significantly decreased the incidence of Antibiotic associated diarrhoea (AAD) and adverse effects related to the use of antibiotics.[56]
- Bacillus subtilis CU1 supplementation effectively and safely stimulate immune responses in humans by increasing the levels of SIgA in saliva and stools.[57]
Safety
In animals
B. subtilis was reviewed by the US FDA Center for Veterinary Medicine and found to present no safety concerns when used in direct-fed microbial products, so the Association of American Feed Control Officials has listed it approved for use as an animal feed ingredient under Section 36.14 "Direct-fed Microorganisms". The Canadian Food Inspection Agency Animal Health and Production Feed Section has classified Bacillus culture dehydrated approved feed ingredients as a silage additive under Schedule IV-Part 2-Class 8.6 and assigned the International Feed Ingredient number IFN 8-19-119.
In humans
B. subtilis spores can survive the extreme heat during cooking. Some B. subtilis strains are responsible for causing ropiness — a sticky, stringy consistency caused by bacterial production of long-chain polysaccharides — in spoiled bread dough. For a long time, bread ropiness was associated uniquely with B. subtilis species by biochemical tests. Molecular assays (randomly amplified polymorphic DNA PCR assay, denaturing gradient gel electrophoresis analysis, and sequencing of the V3 region of 16S ribosomal DNA) revealed greater Bacillus species variety in ropy breads, which all seems to have a positive amylase activity and high heat resistance.[58]
B. subtilis and substances derived from it has been evaluated by different authoritative bodies for their safe and beneficial use in food. In the United States, an opinion letter issued in the early 1960s by the Food and Drug Administration (FDA) recognized some substances derived from microorganisms as Generally recognized as safe (GRAS), including carbohydrase and protease enzymes from B. subtilis. The opinions were predicated on the use of nonpathogenic and nontoxicogenic strains of the respective organisms and on the use of current good manufacturing practices.[59] The FDA stated the enzymes derived from the B. subtilis strain were in common use in food prior to January 1, 1958, and that nontoxigenic and nonpathogenic strains of B. subtilis are widely available and have been safely used in a variety of food applications. This includes consumption of Japanese fermented soy bean, in the form of Natto, which is commonly consumed in Japan, and contains as many as 108 viable cells per gram. The fermented beans are recognized for their contribution to a healthy gut flora and vitamin K2 intake; during this long history of widespread use, natto has not been implicated in adverse events potentially attributable to the presence of B. subtilis. The natto product and the B. subtilis natto as its principal component are FOSHU (Foods for Specified Health Use) approved by the Japanese Ministry of Health, Labour and Welfare as effective for preservation of health.[60]
B. subtilis has been granted "Qualified Presumption of Safety" status by the European Food Safety Authority.[61] B. subtilis is part of the authoritative list of microorganisms with a documented history of safe use in food, established by the International Dairy Federation in collaboration with the European Food and Feed Cultures Association in 2002, and updated in 2012. Bacillus subtilis is Generally Recognized as Safe (GRAS) by the FDA, meaning this bacterium is not harmful to animals or humans. Bacillus subtilis produces 66 anti-microbial substances.[62]
See also
References
- ↑ Euzéby JP (2008). "Bacillus". List of Prokaryotic names with Standing in Nomenclature. Archived from the original on 14 December 2008. Retrieved 2008-11-18.
- ↑ Ambrosiano N (1999-06-30). "Lab biodetector tests to be safe, public to be well informed". Press release. Los Alamos National Labs. Archived from the original on September 21, 2008. Retrieved 2008-11-18.
- ↑ Ehrenberg CG (1835). Physikalische Abhandlungen der Koeniglichen Akademie der Wissenschaften zu Berlin aus den Jahren 1833–1835. pp. 145–336.
- ↑ Cohn, Ferdinand (1872). "Untersuchungen über Bacterien". Beiträge zur Biologie der Pflanzen. 1. pp. 127–224.
- ↑ Yu, Allen Chi-Shing; Loo, Jacky Foo Chuen; Yu, Samuel; Kong, Siu Kai; Chan, Ting-Fung (2013). "Monitoring bacterial growth using tunable resistive pulse sensing with a pore-based technique". Applied microbiology and biotechnology. 98 (2): 855–862. doi:10.1007/s00253-013-5377-9. PMID 24287933.
- ↑ Madigan, M; Martinko, J, eds. (2005). Brock Biology of Microorganisms (11th ed.). Prentice Hall. ISBN 0-13-144329-1.
- ↑ Nakano, Michiko M.; Zuber, Peter (1998). "Anaerobic Growth of A "Strict Aerobe" (Bacillus Subtilis)". Annual Review of Microbiology. 52 (1): 165–90. doi:10.1146/annurev.micro.52.1.165. PMID 9891797.
- ↑ "Isolation and Identification of Pathogenic Bacteria and Fungi from Some Sudanese Banknote Currency". www.medwelljournals.com. Retrieved 2016-12-03.
- ↑ Behera, B.C.; Patra, M.; Dutta, S.K.; Thatoi, H.N. (2014-01-23). "Isolation and Characterisation of Sulphur Oxidising Bacteria from Mangrove Soil of Mahanadi River Delta and Their Sulphur Oxidising Ability". Journal of Applied & Environmental Microbiology. 2 (1). doi:10.12691/jaem-2-1-1.
- ↑ PROKARYOTAE, REGNUM. "ABIS Encyclopedia". www.tgw1916.net. Retrieved 2016-12-03.
- ↑ Leifert, Carlo; Epton, Harold Arthur Stourton; Sigee, David Charles (Sep 30, 1993), Biological control of post-harvest diseases, retrieved 2016-12-03
- ↑ Hong, Huynh A.; Khaneja, Reena; Tam, Nguyen M.K.; Cazzato, Alessia; Tan, Sisareuth; Urdaci, Maria; Brisson, Alain; Gasbarrini, Antonio; Barnes, Ian; Cutting, Simon M. (2009). "Bacillus subtilis isolated from the human gastrointestinal tract". Research in Microbiology. 160 (2): 134–43. doi:10.1016/j.resmic.2008.11.002. PMID 19068230.
- ↑ Noirot P (2007). "Replication of the Bacillus subtilis chromosome". In Graumann P. Bacillus: Cellular and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-12-7.
- ↑ Kobayashi, K.; Ehrlich, S. D.; Albertini, A.; Amati, G.; Andersen, K. K.; Arnaud, M.; Asai, K.; Ashikaga, S.; Aymerich, S.; Bessieres, P.; Boland, F.; Brignell, S. C.; Bron, S.; Bunai, K.; Chapuis, J.; Christiansen, L. C.; Danchin, A.; Debarbouille, M.; Dervyn, E.; Deuerling, E.; Devine, K.; Devine, S. K.; Dreesen, O.; Errington, J.; Fillinger, S.; Foster, S. J.; Fujita, Y.; Galizzi, A.; Gardan, R.; et al. (2003). "Essential Bacillus subtilis genes". Proceedings of the National Academy of Sciences. 100 (8): 4678–83. Bibcode:2003PNAS..100.4678K. doi:10.1073/pnas.0730515100. JSTOR 3144001. PMC 153615
 . PMID 12682299.
. PMID 12682299. - ↑ Yu, Chi-Shing; Yim, Kay-Yuen; Tsui, Stephen Kwok-Wing; Chan, Ting-Fung (2012). "Complete Genome Sequence of Bacillus subtilis Strain QB928, a Strain Widely Used in B. subtilis Genetic Studies". Journal of Bacteriology. 194 (22): 6308–6309. doi:10.1128/JB.01533-12. PMID 23105055.
- ↑ Saito, Shinichi; Kakeshita, Hiroshi; Nakamura, Kouji (2009). "Novel small RNA-encoding genes in the intergenic regions of Bacillus subtilis". Gene. 428 (1–2): 2–8. doi:10.1016/j.gene.2008.09.024. PMID 18948176.
- ↑ Ashlee M. Earl; Richard Losick; Roberto Kolter (June 2008). "Ecology and genomics of Bacillus subtilis". Trends in Microbiology. Elsevier Inc. 16 (6): 269–275. doi:10.1016/j.tim.2008.03.004. ISSN 0966-842X. PMC 2819312
 . PMID 18467096.
. PMID 18467096. - ↑ Saito, Yukiko; Taguchi, Hisataka; Akamatsu, Takashi (2006). "Fate of transforming bacterial genome following incorporation into competent cells of Bacillus subtilis: A continuous length of incorporated DNA". Journal of Bioscience and Bioengineering. 101 (3): 257–62. doi:10.1263/jbb.101.257. PMID 16716928.
- ↑ Saito, Yukiko; Taguchi, Hisataka; Akamatsu, Takashi (2006). "DNA taken into Bacillus subtilis competent cells by lysed-protoplast transformation is not ssDNA but dsDNA". Journal of Bioscience and Bioengineering. 101 (4): 334–9. doi:10.1263/jbb.101.334. PMID 16716942.
- ↑ Akamatsu, Takashi; Taguchi, Hisataka (2001). "Incorporation of the Whole Chromosomal DNA in Protoplast Lysates into Competent Cells of Bacillus subtilis". Bioscience, Biotechnology, and Biochemistry. 65 (4): 823–9. doi:10.1271/bbb.65.823. PMID 11388459.
- ↑ Anagnostopoulos C; Spizizen J (May 1961). "Requirements for transformation in Bacillus subtilis". J. Bacteriol. 81 (5): 741–6. PMC 279084
 . PMID 16561900.
. PMID 16561900. - ↑ Hoelzer MA, Michod RE (June 1991). "DNA repair and the evolution of transformation in Bacillus subtilis. III. Sex with damaged DNA". Genetics. 128 (2): 215–23. PMC 1204460
 . PMID 1906416.
. PMID 1906416. - ↑ Michod RE, Wojciechowski MF, Hoelzer MA (January 1988). "DNA repair and the evolution of transformation in the bacterium Bacillus subtilis". Genetics. 118 (1): 31–9. PMC 1203263
 . PMID 8608929.
. PMID 8608929. - ↑ Wojciechowski MF, Hoelzer MA, Michod RE (March 1989). "DNA repair and the evolution of transformation in Bacillus subtilis. II. Role of inducible repair". Genetics. 121 (3): 411–22. PMC 1203629
 . PMID 2497048.
. PMID 2497048. - ↑ Michod, Richard E.; Bernstein, Harris; Nedelcu, Aurora M. (2008). "Adaptive value of sex in microbial pathogens". Infection, Genetics and Evolution. 8 (3): 267–85. doi:10.1016/j.meegid.2008.01.002. PMID 18295550.
- ↑ Ciprandi G, Scordamaglia A, Venuti D, Caria M, Canonica GW (1986). "In vitro effects of Bacillus subtilis on the immune response". Chemioterapia. 5 (6): 404–7. PMID 3100070.
- ↑ Shylakhovenko, V.A. (June 2003). "Anticancer and Immunostimulatory effects of Nucleoprotein Fraction of Bacillus subtilis". Experimental Oncology. 25: 119–123.
- ↑ Mazza, P. (1994). "The use of Bacillus subtilis as an antidiarrhoeal microorganism". Boll. Chim. Farm. 133 (1): 3–18. PMID 8166962.
- ↑ Horneck G, Klaus DM, Mancinelli RL (March 2010). "Space Microbiology". Microbiology and Molecular Biology Review. 74 (1): 121–156. doi:10.1128/mmbr.00016-09.
- ↑ Dose, K.; Bieger-Dose, A.; Dillmann, R.; Gill, M.; Kerz, O.; Klein, A.; Meinert, H.; Nawroth, T.; Risi, S.; Stridde, C. (1995). "ERA-experiment "space biochemistry"". Advances in Space Research. 16 (8): 119–29. Bibcode:1995AdSpR..16..119D. doi:10.1016/0273-1177(95)00280-R. PMID 11542696.
- ↑ Vaisberg, O.; Fedorov, A.; Dunjushkin, F.; Kozhukhovsky, A.; Smirnov, V.; Avanov, L.; Russell, C.T.; Luhmann, J.G. (1995). "Ion populations in the tail of Venus". Advances in Space Research. 16 (4): 105–18. Bibcode:1995AdSpR..16..105V. doi:10.1016/0273-1177(95)00217-3.
- ↑ Clancy, Paul (Jun 23, 2005). Looking for Life, Searching the Solar System. Cambridge University Press.
- ↑ Horneck, G.; Klaus, D. M.; Mancinelli, R. L. (2010). "Space Microbiology". Microbiology and Molecular Biology Reviews. 74 (1): 121–56. doi:10.1128/MMBR.00016-09. PMC 2832349
 . PMID 20197502.
. PMID 20197502. - ↑ Fajardo-Cavazos, Patricia; Link, Lindsey; Melosh, H. Jay; Nicholson, Wayne L. (2005). "Bacillus subtilisSpores on Artificial Meteorites Survive Hypervelocity Atmospheric Entry: Implications for Lithopanspermia". Astrobiology. 5 (6): 726–36. Bibcode:2005AsBio...5..726F. doi:10.1089/ast.2005.5.726. PMID 16379527.
- ↑ Brandstätter, Franz; Brack, André; Baglioni, Pietro; Cockell, Charles S.; Demets, René; Edwards, Howell G.M.; Kurat, Gero; Osinski, Gordon R.; Pillinger, Judith M.; Roten, Claude-Alain; Sancisi-Frey, Suzy (2008). "Mineralogical alteration of artificial meteorites during atmospheric entry. The STONE-5 experiment". Planetary and Space Science. 56 (7): 976–84. Bibcode:2008P&SS...56..976B. doi:10.1016/j.pss.2007.12.014.
- ↑ Wassmann, Marko; Moeller, Ralf; Rabbow, Elke; Panitz, Corinna; Horneck, Gerda; Reitz, Günther; Douki, Thierry; Cadet, Jean; Stan-Lotter, Helga; Cockell, Charles S.; Rettberg, Petra (2012). "Survival of Spores of the UV-ResistantBacillus subtilisStrain MW01 After Exposure to Low-Earth Orbit and Simulated Martian Conditions: Data from the Space Experiment ADAPT on EXPOSE-E". Astrobiology. 12 (5): 498–507. Bibcode:2012AsBio..12..498W. doi:10.1089/ast.2011.0772. PMID 22680695.
- ↑ Nakamura, L. K. (1989). "Taxonomic Relationship of Black-Pigmented Bacillus subtilis Strains and a Proposal for Bacillus atrophaeus sp. nov". International Journal of Systematic Bacteriology. 39 (3): 295–300. doi:10.1099/00207713-39-3-295.
- ↑ Burke, S. A.; Wright, J. D.; Robinson, M. K.; Bronk, B. V.; Warren, R. L. (2004). "Detection of Molecular Diversity in Bacillus atrophaeus by Amplified Fragment Length Polymorphism Analysis". Applied and Environmental Microbiology. 70 (5): 2786–90. doi:10.1128/AEM.70.5.2786-2790.2004. PMC 404429
 . PMID 15128533.
. PMID 15128533. - ↑ "Project 112/SHAD - Shipboard Hazard and Defense". U.S. Department of Veterans' Affairs. Retrieved 25 February 2015.
- ↑ Gibbons, Henry S.; Broomall, Stacey M.; McNew, Lauren A.; Daligault, Hajnalka; Chapman, Carol; Bruce, David; Karavis, Mark; Krepps, Michael; McGregor, Paul A.; Hong, Charles; Park, Kyong H.; Akmal, Arya; Feldman, Andrew; Lin, Jeffrey S.; Chang, Wenling E.; Higgs, Brandon W.; Demirev, Plamen; Lindquist, John; Liem, Alvin; Fochler, Ed; Read, Timothy D.; Tapia, Roxanne; Johnson, Shannon; Bishop-Lilly, Kimberly A.; Detter, Chris; Han, Cliff; Sozhamannan, Shanmuga; Rosenzweig, C. Nicole; Skowronski, Evan W. (2011). Badger, Jonathan, ed. "Genomic Signatures of Strain Selection and Enhancement in Bacillus atrophaeus var. Globigii, a Historical Biowarfare Simulant". PLoS ONE. 6 (3): e17836. Bibcode:2011PLoSO...617836G. doi:10.1371/journal.pone.0017836. PMC 3064580
 . PMID 21464989.
. PMID 21464989. - ↑ Earl, Ashlee M.; Losick, Richard; Kolter, Roberto (2008). "Ecology and genomics of Bacillus subtilis". Trends in Microbiology. 16 (6): 269–75. doi:10.1016/j.tim.2008.03.004. PMC 2819312
 . PMID 18467096.
. PMID 18467096. - ↑ Jan Maarten van Dijl; Michael Hecker (2013). "Bacillus subtilis: from soil bacterium to super-secreting cell factory". Microbial Cell Factories. 12 (3): 3. doi:10.1186/1475-2859-12-3. Retrieved 15 January 2015.
- ↑ http://www.biopharma.novozymes.com/en/hyaluronic-acid/Frequently-asked-questions/Pages/default.aspx
- ↑ http://www.eppo.int/QUARANTINE/fungi/Monilinia_fructicola/MONIFC_ds.pdf
- ↑ "Biocontrol and other beneficial activities of Bacillus subtilis isolated from cowdung microflora". Microbiological Research. 164: 121–130. doi:10.1016/j.micres.2006.10.009.
- ↑ "Biological control of peach brown rot (Monilinia spp.) by Bacillus subtilis CPA-8 is based on production of fengycin-like lipopeptides". European Journal of Plant Pathology. 132: 609–619. doi:10.1007/s10658-011-9905-0.
- ↑ Sharaf-Eldin, Mahmoud; Elkholy, Shereen; Fernández, José-Antonio; Junge, Helmut; Cheetham, Ronald; Guardiola, José; Weathers, Pamela (2008). "Bacillus subtilisFZB24® Affects Flower Quantity and Quality of Saffron (Crocus sativus)". Planta Medica. 74 (10): 1316–20. doi:10.1055/s-2008-1081293. PMC 3947403
 . PMID 18622904.
. PMID 18622904. - ↑ Harrigan GG, Ridley WP, Miller KD, et al. (October 2009). "The forage and grain of MON 87460, a drought-tolerant corn hybrid, are compositionally equivalent to that of conventional corn". J. Agric. Food Chem. 57 (20): 9754–63. doi:10.1021/jf9021515. PMID 19778059.
- ↑ USDA: Determination of Nonregulated Status for MON 87460 Corn (Zea mays L)
- ↑ "The International Pharmacopoeia Fourth Edition - Pharmacopoea Internationalis Editio Quarta - Fourth Supplement: Methods of Analysis: 5. Pharmaceutical technical procedures: 5.8 Methods of sterilization".
- ↑ "Andersen Products: AN-2203 Biological Indicator for EO (25/box)".
- ↑ Yu, Allen Chi-Shing; Yim, Aldrin Kay-Yuen; Mat, Wai-Kin; Tong, Amy Hin-Yan; Lok, Si; Xue, Hong; Tsui, Stephen Kwok-Wing; Wong, Jeffrey Tsz-Fei; Chan, Ting-Fung (2014). "Mutations Enabling Displacement of Tryptophan by 4-Fluorotryptophan as a Canonical Amino Acid of the Genetic Code". Genome Biology and Evolution. 6 (3): 629–641. doi:10.1093/gbe/evu044. PMID 24572018.
- ↑ Hosoi, T; Ametani, A; Kiuchi, K; Kaminogawa, S (October 2000). "Improved growth and viability of lactobacilli in the presence of Bacillus subtilis (natto), catalase, or subtilisin.". Canadian journal of microbiology. 46 (10): 892–7. PMID 11068675.
- ↑ Green, DH; Wakeley, PR; Page, A; Barnes, A; Baccigalupi, L; Ricca, E; Cutting, SM (September 1999). "Characterization of Two Bacillus Probiotics". Appl Environ Microbiol. 65 (9): 4288–4291.
- ↑ Angioi, A (1995). "Adhesiveness of Bacillus subtilis Strains to Epithelial Cells Cultured in vitro". Microbial Ecology in Health and Disease. 8 (2): 71–77. doi:10.3109/08910609509141385.
- ↑ Horosheva, Tatiana V.; Vodyanoy, Vitaly; Sorokulova, Iryna. "Efficacy of Bacillus probiotics in prevention of antibiotic‐associated diarrhoea: a randomized, double‐blind, placebo‐controlled clinical trial". JMM Case Reports: 1–6. doi:10.1099/jmmcr.0.004036.
- ↑ Lefevre, Marie; Racedo, Silvia; Ripert, Gabrielle; Housez, Béatrice; Cazaubiel, Murielle; Maudet, Corinne; Jüsten, Peter; Marteau, Philippe; Urdaci, Maria (2015). "Probiotic strain Bacillus subtilis CU1 stimulates immune system of elderly during common infectious disease period: a randomized, double-blind placebo-controlled study". Immun Ageing. 12 (24): 1–11. doi:10.1186/s12979-015-0051-y. PMC 4669646
 . PMID 26640504.
. PMID 26640504. - ↑ Pepe O, Blaiotta G, Moschetti G, Greco T, Villani F (April 2003). "Rope-producing strains of Bacillus spp. from wheat bread and strategy for their control by lactic acid bacteria". Appl. Environ. Microbiol. 69 (4): 2321–9. doi:10.1128/AEM.69.4.2321-2329.2003. PMC 154770
 . PMID 12676716.
. PMID 12676716. - ↑ "FDA partial list of microorganisms". Food and Drug Administration. 2002.
- ↑ Gibson G.R., 2005. Functional Foods: Perspectives on foods for specific health uses (FOSHU), Colette Shortt, Yakult UK. Volume 1, page 7-1.
- ↑ EFSA Panel on Biological Hazards (BIOHAZ) (2010). "Scientific opinion on the maintenance of the list of QPS microorganisms intentionally added to food or feed (2010 update)". EFSA Journal. 8 (12): 1944.
- ↑ Olmos and Paniagua-Michel, J Microb Biochem Technol 2014, 6:7:361-365
External links
![]() Media related to Bacillus subtilis at Wikimedia Commons
Media related to Bacillus subtilis at Wikimedia Commons
- SubtiWiki "up-to-date information for all genes of Bacillus subtilis"
- Bacillus subtilis Final Risk Assessment on EPA.gov
- Bacillus subtilis genome browser
- Type strain of Bacillus subtilis at BacDive - the Bacterial Diversity Metadatabase